LuxG is a functioning flavin reductase for bacterial luminescence
- PMID: 18156264
- PMCID: PMC2258676
- DOI: 10.1128/JB.01660-07
LuxG is a functioning flavin reductase for bacterial luminescence
Abstract
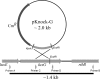
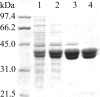
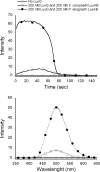
The luxG gene is part of the lux operon of marine luminous bacteria. luxG has been proposed to be a flavin reductase that supplies reduced flavin mononucleotide (FMN) for bacterial luminescence. However, this role has never been established because the gene product has not been successfully expressed and characterized. In this study, luxG from Photobacterium leiognathi TH1 was cloned and expressed in Escherichia coli in both native and C-terminal His6-tagged forms. Sequence analysis indicates that the protein consists of 237 amino acids, corresponding to a subunit molecular mass of 26.3 kDa. Both expressed forms of LuxG were purified to homogeneity, and their biochemical properties were characterized. Purified LuxG is homodimeric and has no bound prosthetic group. The enzyme can catalyze oxidation of NADH in the presence of free flavin, indicating that it can function as a flavin reductase in luminous bacteria. NADPH can also be used as a reducing substrate for the LuxG reaction, but with much less efficiency than NADH. With NADH and FMN as substrates, a Lineweaver-Burk plot revealed a series of convergent lines characteristic of a ternary-complex kinetic model. From steady-state kinetics data at 4 degrees C pH 8.0, Km for NADH, Km for FMN, and kcat were calculated to be 15.1 microM, 2.7 microM, and 1.7 s(-1), respectively. Coupled assays between LuxG and luciferases from P. leiognathi TH1 and Vibrio campbellii also showed that LuxG could supply FMNH- for light emission in vitro. A luxG gene knockout mutant of P. leiognathi TH1 exhibited a much dimmer luminescent phenotype compared to the native P. leiognathi TH1, implying that LuxG is the most significant source of FMNH- for the luminescence reaction in vivo.
Figures




References
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26824-826, 828. - PubMed
-
- Andrews, S. C., D. Shipley, J. N. Keen, J. B. Findlay, P. M. Harrison, and J. R. Guest. 1992. The hemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 302247-252. - PubMed
-
- Bruns, C. M., and P. A. Karplus. 1995. Refined crystal structure of spinach ferredoxin reductase at 1.7 Å resolution: oxidized, reduced and 2′-phospho-5′-AMP bound states. J. Mol. Biol. 247125-145. - PubMed
-
- Chaiyen, P., C. Suadee, and P. Wilairat. 2001. A novel two-protein component flavoprotein hydroxylase. Eur. J. Biochem. 2685550-5561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

