Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants
- PMID: 18156295
- PMCID: PMC2245839
- DOI: 10.1104/pp.107.111393
Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants
Abstract
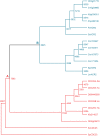
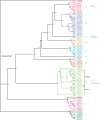
In addition to the major serine/threonine-specific phosphoprotein phosphatase, Mg(2+)-dependent phosphoprotein phosphatase, and protein tyrosine phosphatase families, there are novel protein phosphatases, including enzymes with aspartic acid-based catalysis and subfamilies of protein tyrosine phosphatases, whose evolutionary history and representation in plants is poorly characterized. We have searched the protein data sets encoded by the well-finished nuclear genomes of the higher plants Arabidopsis (Arabidopsis thaliana) and Oryza sativa, and the latest draft data sets from the tree Populus trichocarpa and the green algae Chlamydomonas reinhardtii and Ostreococcus tauri, for homologs to several classes of novel protein phosphatases. The Arabidopsis proteins, in combination with previously published data, provide a complete inventory of known types of protein phosphatases in this organism. Phylogenetic analysis of these proteins reveals a pattern of evolution where a diverse set of protein phosphatases was present early in the history of eukaryotes, and the division of plant and animal evolution resulted in two distinct sets of protein phosphatases. The green algae occupy an intermediate position, and show similarity to both plants and animals, depending on the protein. Of specific interest are the lack of cell division cycle (CDC) phosphatases CDC25 and CDC14, and the seeming adaptation of CDC14 as a protein interaction domain in higher plants. In addition, there is a dramatic increase in proteins containing RNA polymerase C-terminal domain phosphatase-like catalytic domains in the higher plants. Expression analysis of Arabidopsis phosphatase genes differentially amplified in plants (specifically the C-terminal domain phosphatase-like phosphatases) shows patterns of tissue-specific expression with a statistically significant number of correlated genes encoding putative signal transduction proteins.
Figures

References
-
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T (2004) Protein tyrosine phosphatases in the human genome. Cell 117 699–711 - PubMed
-
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14 48–54 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

