Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation
- PMID: 18157086
- PMCID: PMC2168397
- DOI: 10.1038/sj.emboj.7601967
Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation
Abstract
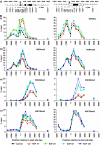
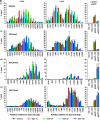
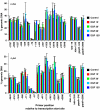
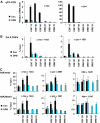
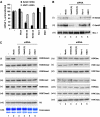
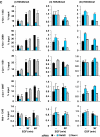
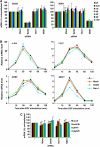
Understanding the function of histone modifications across inducible genes in mammalian cells requires quantitative, comparative analysis of their fate during gene activation and identification of enzymes responsible. We produced high-resolution comparative maps of the distribution and dynamics of H3K4me3, H3K36me3, H3K79me2 and H3K9ac across c-fos and c-jun upon gene induction in murine fibroblasts. In unstimulated cells, continuous turnover of H3K9 acetylation occurs on all K4-trimethylated histone H3 tails; distribution of both modifications coincides across promoter and 5' part of the coding region. In contrast, K36- and K79-methylated H3 tails, which are not dynamically acetylated, are restricted to the coding regions of these genes. Upon stimulation, transcription-dependent increases in H3K4 and H3K36 trimethylation are seen across coding regions, peaking at 5' and 3' ends, respectively. Addressing molecular mechanisms involved, we find that Huntingtin-interacting protein HYPB/Setd2 is responsible for virtually all global and transcription-dependent H3K36 trimethylation, but not H3K36-mono- or dimethylation, in these cells. These studies reveal four distinct layers of histone modification across inducible mammalian genes and show that HYPB/Setd2 is responsible for H3K36 trimethylation throughout the mouse nucleus.
Figures


References
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, Macalpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schübeler D (2007) Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J (advance online publication, 15 November 2007, doi:10.1038/sj.emboj.7601926) - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

