Functional inhibition related to structure of a highly potent insulin-specific CD8 T cell clone using altered peptide ligands
- PMID: 18157812
- PMCID: PMC2901522
- DOI: 10.1002/eji.200737762
Functional inhibition related to structure of a highly potent insulin-specific CD8 T cell clone using altered peptide ligands
Abstract
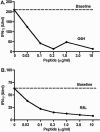
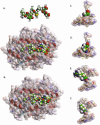
Insulin-reactive CD8 T cells are amongst the earliest islet-infiltrating CD8 T cells in NOD mice. Cloned insulin B15-23-reactive cells (designated G9C8), restricted by H-2K(d), are highly diabetogenic. We used altered peptide ligands (APL) substituted at TCR contact sites, positions (p)6 and 8, to investigate G9C8 T cell function and correlated this with structure. Cytotoxicity and IFN-gamma production assays revealed that p6G and p8R could not be replaced by any naturally occurring amino acid without abrogating recognition and functional response by the G9C8 clone. When tested for antagonist activity with APL differing from the native peptide at either of these positions, the peptide variants, G6H and R8L showed the capacity to reduce the agonist response to the native peptide. The antagonist activity in cytotoxicity and IFN-gamma production assays can be correlated with conformational changes induced by different structures of the MHC-peptide complexes, shown by molecular modeling. We conclude that p6 and p8 of the insulin B15-23 peptide are very important for TCR stimulation of this clone and no substitutions are tolerated at these positions in the peptide. This is important in considering the therapeutic use of peptides as APL that encompass both CD4 and CD8 epitopes of insulin.
Figures



References
-
- Evavold BD, Sloan-Lancaster J, Allen PM. Tickling the TCR: selective T-cell functions stimulated by altered peptide ligands. Immunol. Today. 1993;14:602–609. - PubMed
-
- Sloan-Lancaster J, Allen PM. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996;14:1–27. - PubMed
-
- Kalergis AM, Nathenson SG. Altered peptide ligand-mediated TCR antagonism can be modulated by a change in a single amino acid residue within the CDR3 beta of an MHC class I-restricted TCR. J. Immunol. 2000;165:280–285. - PubMed
-
- Ford ML, Evavold BD. An MHC anchor-substituted analog of myelin oligodendrocyte glycoprotein 35–55 induces IFN-gamma and autoantibodies in the absence of experimental autoimmune encephalomyelitis and optic neuritis. Eur. J. Immunol. 2004;34:388–397. - PubMed
-
- Ryan KR, McNeil LK, Dao C, Jensen PE, Evavold BD. Modification of peptide interaction with MHC creates TCR partial agonists. Cell. Immunol. 2004;227:70–78. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

