Mycobacterial phenolic glycolipid virulence factor biosynthesis: mechanism and small-molecule inhibition of polyketide chain initiation
- PMID: 18158259
- PMCID: PMC2276623
- DOI: 10.1016/j.chembiol.2007.11.010
Mycobacterial phenolic glycolipid virulence factor biosynthesis: mechanism and small-molecule inhibition of polyketide chain initiation
Abstract
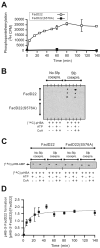
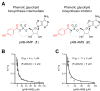
Phenolic glycolipids (PGLs) are polyketide-derived virulence factors produced by Mycobacterium tuberculosis, M. leprae, and other mycobacterial pathogens. We have combined bioinformatic, genetic, biochemical, and chemical biology approaches to illuminate the mechanism of chain initiation required for assembly of the p-hydroxyphenyl-polyketide moiety of PGLs. Our studies have led to the identification of a stand-alone, didomain initiation module, FadD22, comprised of a p-hydroxybenzoic acid adenylation domain and an aroyl carrier protein domain. FadD22 forms an acyl-S-enzyme covalent intermediate in the p-hydroxyphenyl-polyketide chain assembly line. We also used this information to develop a small-molecule inhibitor of PGL biosynthesis. Overall, these studies provide insights into the biosynthesis of an important group of small-molecule mycobacterial virulence factors and support the feasibility of targeting PGL biosynthesis to develop new drugs to treat mycobacterial infections.
Figures

Similar articles
-
Cooperation between a coenzyme A-independent stand-alone initiation module and an iterative type I polyketide synthase during synthesis of mycobacterial phenolic glycolipids.J Am Chem Soc. 2009 Nov 25;131(46):16744-50. doi: 10.1021/ja904792q. J Am Chem Soc. 2009. PMID: 19799378 Free PMC article.
-
Biosynthesis of cell envelope-associated phenolic glycolipids in Mycobacterium marinum.J Bacteriol. 2015 Mar;197(6):1040-50. doi: 10.1128/JB.02546-14. Epub 2015 Jan 5. J Bacteriol. 2015. PMID: 25561717 Free PMC article.
-
Mycobacterial phenolic glycolipid synthesis is regulated by cAMP-dependent lysine acylation of FadD22.Microbiology (Reading). 2017 Mar;163(3):373-382. doi: 10.1099/mic.0.000440. Epub 2017 Mar 29. Microbiology (Reading). 2017. PMID: 28141495
-
The dimycocerosate ester polyketide virulence factors of mycobacteria.Prog Lipid Res. 2005 Sep;44(5):259-302. doi: 10.1016/j.plipres.2005.07.001. Epub 2005 Aug 3. Prog Lipid Res. 2005. PMID: 16115688 Review.
-
The Emergence of Phenolic Glycans as Virulence Factors in Mycobacterium tuberculosis.ACS Chem Biol. 2017 Aug 18;12(8):1969-1979. doi: 10.1021/acschembio.7b00394. Epub 2017 Jul 19. ACS Chem Biol. 2017. PMID: 28692249 Review.
Cited by
-
Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family.BMC Microbiol. 2012 Jun 22;12:118. doi: 10.1186/1471-2180-12-118. BMC Microbiol. 2012. PMID: 22726990 Free PMC article.
-
Active site-directed proteomic probes for adenylation domains in nonribosomal peptide synthetases.Chem Commun (Camb). 2015 Feb 11;51(12):2262-5. doi: 10.1039/c4cc09412c. Chem Commun (Camb). 2015. PMID: 25563804 Free PMC article.
-
Inactivation of tesA reduces cell wall lipid production and increases drug susceptibility in mycobacteria.J Biol Chem. 2011 Jul 15;286(28):24616-25. doi: 10.1074/jbc.M111.247601. Epub 2011 May 18. J Biol Chem. 2011. PMID: 21592957 Free PMC article.
-
Mycobacterial Phenolic Glycolipid Triggers ATP-Mediated Neuronal P2X3 Signaling and Cough.bioRxiv [Preprint]. 2025 May 6:2025.05.01.651726. doi: 10.1101/2025.05.01.651726. bioRxiv. 2025. PMID: 40654771 Free PMC article. Preprint.
-
Targeting the mycobacterial envelope for tuberculosis drug development.Expert Rev Anti Infect Ther. 2012 Sep;10(9):1023-36. doi: 10.1586/eri.12.91. Expert Rev Anti Infect Ther. 2012. PMID: 23106277 Free PMC article. Review.
References
-
- World Health Organization. Tuberculosis. 2007. http://www.who.int/mediacentre/factsheets/fs104/en/print.html.
-
- World Health Organization. Leprosy. 2007. http://www.who.int/mediacentre/factsheets/fs101/en/
-
- Aziz MA, Wright A, Laszlo A, De Muynck A, Portaels F, Van Deun A, Wells C, Nunn P, Blanc L, Raviglione M. Epidemiology of antituberculosis drug resistance (the Global Project on Anti-tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet. 2006;368:2142–2154. - PubMed
-
- Centers for Disease Control and Prevention. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs--worldwide, 2000-2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. - PubMed
-
- Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363:1209–1219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

