Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles
- PMID: 18158335
- PMCID: PMC2373489
- DOI: 10.1083/jcb.200710061
Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles
Abstract
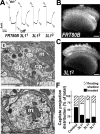
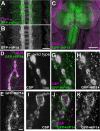
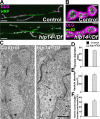
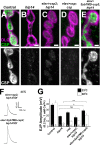
Posttranslational modification through palmitoylation regulates protein localization and function. In this study, we identify a role for the Drosophila melanogaster palmitoyl transferase Huntingtin-interacting protein 14 (HIP14) in neurotransmitter release. hip14 mutants show exocytic defects at low frequency stimulation and a nearly complete loss of synaptic transmission at higher temperature. Interestingly, two exocytic components known to be palmitoylated, cysteine string protein (CSP) and SNAP25, are severely mislocalized at hip14 mutant synapses. Complementary DNA rescue and localization experiments indicate that HIP14 is required solely in the nervous system and is essential for presynaptic function. Biochemical studies indicate that HIP14 palmitoylates CSP and that CSP is not palmitoylated in hip14 mutants. Furthermore, the hip14 exocytic defects can be suppressed by targeting CSP to synaptic vesicles using a chimeric protein approach. Our data indicate that HIP14 controls neurotransmitter release by regulating the trafficking of CSP to synapses.
Figures



References
-
- Arnold, C., N. Reisch, C. Leibold, S. Becker, K. Prufert, K. Sautter, D. Palm, S. Jatzke, S. Buchner, and E. Buchner. 2004. Structure-function analysis of the cysteine string protein in Drosophila: cysteine string, linker and C terminus. J. Exp. Biol. 207:1323–1334. - PubMed
-
- Babu, P., R.J. Deschenes, and L.C. Robinson. 2004. Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J. Biol. Chem. 279:27138–27147. - PubMed
-
- Bronk, P., J.J. Wenniger, K. Dawson-Scully, X. Guo, S. Hong, H.L. Atwood, and K.E. Zinsmaier. 2001. Drosophila Hsc70-4 is critical for neurotransmitter exocytosis in vivo. Neuron. 30:475–488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

