Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin
- PMID: 18158898
- PMCID: PMC2610362
- DOI: 10.1016/j.molcel.2007.12.002
Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin
Abstract
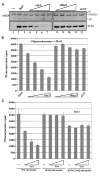
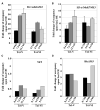
Dot1 (Disruptor of telomeric silencing-1) is a histone H3 lysine 79 methyltransferase that contributes to the establishment of heterochromatin boundary and has been linked to transcription elongation. We found that histone H4 N-terminal domain, unlike other histone tails, interacts with Dot1 and is essential for H3 K79 methylation. Furthermore, we show that the heterochromatin protein Sir3 inhibits Dot1-mediated methylation and that this inhibition is dependent on lysine 16 of H4. Sir3 and Dot1 bind the same short basic patch of histone H4 tail, and Sir3 also associates with the residues surrounding H3 K79 in a methylation-sensitive manner. Thus, Sir3 and Dot1 compete for the same molecular target on chromatin. ChIP analyses support a model in which acetylation of H4 lysine 16 displaces Sir3, allowing Dot1 to bind and methylate H3 lysine 79, which in turn further blocks Sir3 binding/spreading. This draws a detailed picture of the succession of molecular events occurring during the establishment of telomeric heterochromatin boundaries.
Figures




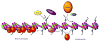
References
-
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. - PubMed
-
- Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem. 2002;277:4778–4781. - PubMed
-
- Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

