Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair
- PMID: 18158901
- PMCID: PMC2275782
- DOI: 10.1016/j.molcel.2007.12.005
Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair
Abstract
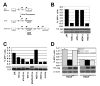
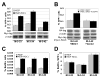
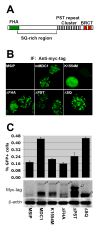
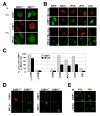
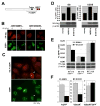
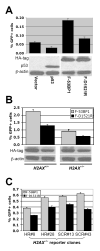
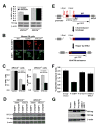
Phosphorylated histone H2AX ("gamma-H2AX") recruits MDC1, 53BP1, and BRCA1 to chromatin near a double-strand break (DSB) and facilitates efficient repair of the break. It is unclear to what extent gamma-H2AX-associated proteins act in concert and to what extent their functions within gamma-H2AX chromatin are distinct. We addressed this question by comparing the mechanisms of action of MDC1 and 53BP1 in DSB repair (DSBR). We find that MDC1 functions primarily in homologous recombination/sister chromatid recombination, in a manner strictly dependent upon its ability to interact with gamma-H2AX but, unexpectedly, not requiring recruitment of 53BP1 or BRCA1 to gamma-H2AX chromatin. In contrast, 53BP1 functions in XRCC4-dependent nonhomologous end-joining, likely mediated by its interaction with dimethylated lysine 20 of histone H4 but, surprisingly, independent of H2AX. These results suggest a specialized adaptation of the "histone code" in which distinct histone tail-protein interactions promote engagement of distinct DSBR pathways.
Figures
References
-
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. - PubMed
-
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

