Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals
- PMID: 18159944
- PMCID: PMC2151084
- DOI: 10.1371/journal.ppat.0030197
Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals
Abstract
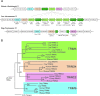
TRIM5alpha provides a cytoplasmic block to retroviral infection, and orthologs encoded by some primates are active against HIV. Here, we present an evolutionary comparison of the TRIM5 gene to its closest human paralogs: TRIM22, TRIM34, and TRIM6. We show that TRIM5 and TRIM22 have a dynamic history of gene expansion and loss during the evolution of mammals. The cow genome contains an expanded cluster of TRIM5 genes and no TRIM22 gene, while the dog genome encodes TRIM22 but has lost TRIM5. In contrast, TRIM6 and TRIM34 have been strictly preserved as single gene orthologs in human, dog, and cow. A more focused analysis of primates reveals that, while TRIM6 and TRIM34 have evolved under purifying selection, TRIM22 has evolved under positive selection as was previously observed for TRIM5. Based on TRIM22 sequences obtained from 27 primate genomes, we find that the positive selection of TRIM22 has occurred episodically for approximately 23 million years, perhaps reflecting the changing pathogenic landscape. However, we find that the evolutionary episodes of positive selection that have acted on TRIM5 and TRIM22 are mutually exclusive, with generally only one of these genes being positively selected in any given primate lineage. We interpret this to mean that the positive selection of one gene has constrained the adaptive flexibility of its neighbor, probably due to genetic linkage. Finally, we find a striking congruence in the positions of amino acid residues found to be under positive selection in both TRIM5alpha and TRIM22, which in both proteins fall predominantly in the beta2-beta3 surface loop of the B30.2 domain. Astonishingly, this same loop is under positive selection in the multiple cow TRIM5 genes as well, indicating that this small structural loop may be a viral recognition motif spanning a hundred million years of mammalian evolution.
Conflict of interest statement
Figures


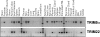

References
-
- Malim MH. Natural resistance to HIV infection: the Vif–APOBEC interaction. C R Biol. 2006;329:871–875. - PubMed
-
- Perez O, Hope TJ. Cellular restriction factors affecting the early stages of HIV replication. Curr HIV/AIDS Rep. 2006;3:20–25. - PubMed
-
- Newman RM, Johnson WE. A brief history of TRIM5alpha. AIDS Rev. 2007;9:114–125. - PubMed
-
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

