Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma
- PMID: 18161048
- PMCID: PMC2865182
- DOI: 10.1002/hep.22110
Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma
Abstract
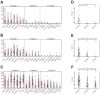
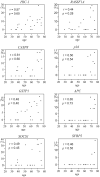
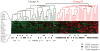
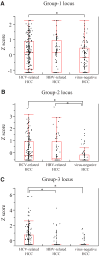
Aberrant DNA methylation is an important epigenetic alteration in hepatocellular carcinoma (HCC). However, the molecular processes underlying the methylator phenotype and the contribution of hepatitis viruses are poorly understood. The current study is a comprehensive methylation analysis of human liver tissue specimens. A total of 176 liver tissues, including 77 pairs of HCCs and matching noncancerous liver and 22 normal livers, were analyzed for methylation. Methylation of 19 epigenetic markers was quantified, and the results were correlated with different disease states and the presence or absence of hepatitis B virus (HBV) and hepatitis C virus (HCV) infections. Based on methylation profiles, the 19 loci were categorized into 3 groups. Normal liver tissues showed methylation primarily in group 1 loci (HIC-1, CASP8, GSTP1, SOCS1, RASSF1A, p16, APC), which was significantly higher than group 2 (CDH1, RUNX3, RIZ1, SFRP2, MINT31) and group 3 markers (COX2, MINT1, CACNA1G, RASSF2, MINT2, Reprimo, DCC) (P < 0.0001). Noncancerous livers demonstrated increased methylation in both group 1 and group 2 loci. Methylation was significantly more abundant in HCV-positive livers compared with normal liver tissues. Conversely, HCC showed frequent methylation at each locus investigated in all 3 groups. However, the group 3 loci showed more dense and frequent methylation in HCV-positive cancers compared with both HBV-positive cancers and virus-negative cancers (P < 0.0001).
Conclusion: Methylation in HCC is frequent but occurs in a gene-specific and disease-specific manner. Methylation profiling allowed us to determine that aberrant methylation is commonly present in normal aging livers, and sequentially progresses with advancing stages of chronic viral infection. Finally, our data provide evidence that HCV infection may accelerate the methylation process and suggests a continuum of increasing methylation with persistent viral infection and carcinogenesis in the liver.
Conflict of interest statement
Potential conflict of interest: Nothing to report.
Figures

References
-
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. - PubMed
-
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. - PubMed
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
