Single-molecule tracking of mRNA exiting from RNA polymerase II
- PMID: 18162559
- PMCID: PMC2224174
- DOI: 10.1073/pnas.0703815105
Single-molecule tracking of mRNA exiting from RNA polymerase II
Abstract
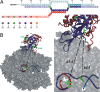
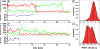
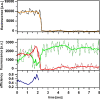
Single-pair fluorescence resonance energy transfer was used to track RNA exiting from RNA polymerase II (Pol II) in elongation complexes. Measuring the distance between the RNA 5' end and three known locations within the elongation complex allows us determine its position by means of triangulation. RNA leaves the polymerase active center cleft via the previously proposed exit tunnel and then disengages from the enzyme surface. When the RNA reaches lengths of 26 and 29 nt, its 5' end associates with Pol II at the base of the dock domain. Because the initiation factor TFIIB binds to the dock domain and exit tunnel, exiting RNA may prevent TFIIB reassociation during elongation. RNA further extends toward the linker connecting to the polymerase C-terminal repeat domain (CTD), which binds the 5'-capping enzyme and other RNA processing factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863–1876. - PubMed
-
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. J Biol Chem. 2005;280:7131–7134. - PubMed
-
- Kettenberger H, Armache KJ, Cramer P. Mol Cell. 2004;16:955–965. - PubMed
-
- Westover KD, Bushnell DA, Kornberg RD. Cell. 2004;119:481–489. - PubMed
-
- Armache KJ, Kettenberger H, Cramer P. Curr Opin Struct Biol. 2005;15:197–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

