HsdR subunit of the type I restriction-modification enzyme EcoR124I: biophysical characterisation and structural modelling
- PMID: 18164032
- PMCID: PMC2878639
- DOI: 10.1016/j.jmb.2007.11.024
HsdR subunit of the type I restriction-modification enzyme EcoR124I: biophysical characterisation and structural modelling
Erratum in
-
Corrigendum to "HsdR Subunit of the Type I Restriction-Modification Enzyme EcoR124I: Biophysical Characterisation and Structural Modelling" [J. Mol. Biol. 376(2) 2008 Feb 15: 438-452.].J Mol Biol. 2020 Mar 27;432(7):2444. doi: 10.1016/j.jmb.2020.02.027. Epub 2020 Mar 18. J Mol Biol. 2020. PMID: 32199672 Free PMC article. No abstract available.
Abstract
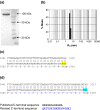
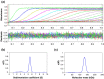
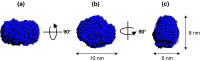
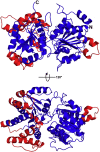
Type I restriction-modification (RM) systems are large, multifunctional enzymes composed of three different subunits. HsdS and HsdM form a complex in which HsdS recognizes the target DNA sequence, and HsdM carries out methylation of adenosine residues. The HsdR subunit, when associated with the HsdS-HsdM complex, translocates DNA in an ATP-dependent process and cleaves unmethylated DNA at a distance of several thousand base-pairs from the recognition site. The molecular mechanism by which these enzymes translocate the DNA is not fully understood, in part because of the absence of crystal structures. To date, crystal structures have been determined for the individual HsdS and HsdM subunits and models have been built for the HsdM-HsdS complex with the DNA. However, no structure is available for the HsdR subunit. In this work, the gene coding for the HsdR subunit of EcoR124I was re-sequenced, which showed that there was an error in the published sequence. This changed the position of the stop codon and altered the last 17 amino acid residues of the protein sequence. An improved purification procedure was developed to enable HsdR to be purified efficiently for biophysical and structural analysis. Analytical ultracentrifugation shows that HsdR is monomeric in solution, and the frictional ratio of 1.21 indicates that the subunit is globular and fairly compact. Small angle neutron-scattering of the HsdR subunit indicates a radius of gyration of 3.4 nm and a maximum dimension of 10 nm. We constructed a model of the HsdR using protein fold-recognition and homology modelling to model individual domains, and small-angle neutron scattering data as restraints to combine them into a single molecule. The model reveals an ellipsoidal shape of the enzymatic core comprising the N-terminal and central domains, and suggests conformational heterogeneity of the C-terminal region implicated in binding of HsdR to the HsdS-HsdM complex.
Figures







Similar articles
-
pHluorin-assisted expression, purification, crystallization and X-ray diffraction data analysis of the C-terminal domain of the HsdR subunit of the Escherichia coli type I restriction-modification system EcoR124I.Acta Crystallogr F Struct Biol Commun. 2016 Sep;72(Pt 9):672-6. doi: 10.1107/S2053230X16011626. Epub 2016 Aug 9. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 27599856 Free PMC article.
-
The DNA recognition subunit of the type IB restriction-modification enzyme EcoAI tolerates circular permutions of its polypeptide chain.J Mol Biol. 1998 Dec 11;284(4):937-48. doi: 10.1006/jmbi.1998.2250. J Mol Biol. 1998. PMID: 9837717
-
Domain structure and subunit interactions in the type I DNA methyltransferase M.EcoR124I.J Mol Biol. 2001 Nov 16;314(1):41-50. doi: 10.1006/jmbi.2001.5123. J Mol Biol. 2001. PMID: 11724530
-
Characterization of an EcoR124I restriction-modification enzyme produced from a deleted form of the DNA-binding subunit, which results in a novel DNA specificity.Folia Microbiol (Praha). 2003;48(3):319-28. doi: 10.1007/BF02931361. Folia Microbiol (Praha). 2003. PMID: 12879741
-
A novel mutant of the type I restriction-modification enzyme EcoR124I is altered at a key stage of the subunit assembly pathway.J Mol Biol. 2000 Dec 1;304(3):301-10. doi: 10.1006/jmbi.2000.4219. J Mol Biol. 2000. PMID: 11090275
Cited by
-
Genomic and Proteomic Characterization of Bacteriophage BH1 Spontaneously Released from Probiotic Lactobacillus rhamnosus Pen.Viruses. 2019 Dec 16;11(12):1163. doi: 10.3390/v11121163. Viruses. 2019. PMID: 31888239 Free PMC article.
-
Adaptive loss of tRNA gene expression leads to phage resistance in a marine Synechococcus cyanobacterium.Nat Microbiol. 2025 Jan;10(1):66-76. doi: 10.1038/s41564-024-01877-6. Epub 2025 Jan 3. Nat Microbiol. 2025. PMID: 39753669 Free PMC article.
-
The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein.Nucleic Acids Res. 2009 Feb;37(3):762-70. doi: 10.1093/nar/gkn988. Epub 2008 Dec 11. Nucleic Acids Res. 2009. PMID: 19074193 Free PMC article.
-
pHluorin-assisted expression, purification, crystallization and X-ray diffraction data analysis of the C-terminal domain of the HsdR subunit of the Escherichia coli type I restriction-modification system EcoR124I.Acta Crystallogr F Struct Biol Commun. 2016 Sep;72(Pt 9):672-6. doi: 10.1107/S2053230X16011626. Epub 2016 Aug 9. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 27599856 Free PMC article.
-
An Mrr-family nuclease motif in the single polypeptide restriction-modification enzyme LlaGI.Nucleic Acids Res. 2009 Nov;37(21):7231-8. doi: 10.1093/nar/gkp795. Nucleic Acids Res. 2009. PMID: 19793866 Free PMC article.
References
-
- Sistla S., Rao D.N. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

