Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation
- PMID: 18166085
- PMCID: PMC2323313
- DOI: 10.1371/journal.pgen.0030235
Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation
Abstract
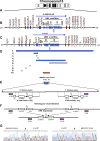
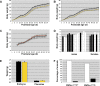
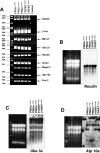
Prader-Willi syndrome (PWS [MIM 176270]) is a neurogenetic disorder characterized by decreased fetal activity, muscular hypotonia, failure to thrive, short stature, obesity, mental retardation, and hypogonadotropic hypogonadism. It is caused by the loss of function of one or more imprinted, paternally expressed genes on the proximal long arm of chromosome 15. Several potential PWS mouse models involving the orthologous region on chromosome 7C exist. Based on the analysis of deletions in the mouse and gene expression in PWS patients with chromosomal translocations, a critical region (PWScr) for neonatal lethality, failure to thrive, and growth retardation was narrowed to the locus containing a cluster of neuronally expressed MBII-85 small nucleolar RNA (snoRNA) genes. Here, we report the deletion of PWScr. Mice carrying the maternally inherited allele (PWScr(m-/p+)) are indistinguishable from wild-type littermates. All those with the paternally inherited allele (PWScr(m+/p-)) consistently display postnatal growth retardation, with about 15% postnatal lethality in C57BL/6, but not FVB/N crosses. This is the first example in a multicellular organism of genetic deletion of a C/D box snoRNA gene resulting in a pronounced phenotype.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




References
-
- Kantor B, Shemer R, Razin A. The Prader-Willi/Angelman imprinted domain and its control center. Cytogenet Genome Res. 2006;113:300–305. - PubMed
-
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. - PubMed
-
- Burd L, Vesely B, Martsolf J, Kerbeshian J. Prevalence study of Prader-Willi syndrome in North Dakota. Am J Med Genet. 1990;37:97–99. - PubMed
-
- Wattendorf DJ, Muenke M. Prader-Willi syndrome. Am Fam Physician. 2005;72:827–830. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

