Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules
- PMID: 18166649
- PMCID: PMC2373496
- DOI: 10.1083/jcb.200705163
Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules
Abstract
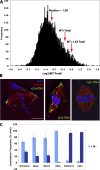
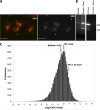
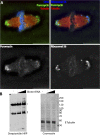
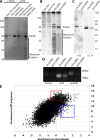
RNA localization is of critical importance in many fundamental cell biological and developmental processes by regulating the spatial control of gene expression. To investigate how spindle-localized RNAs might influence mitosis, we comprehensively surveyed all messenger RNAs (mRNAs) that bound to microtubules during metaphase in both Xenopus laevis egg extracts and mitotic human cell extracts. We identify conserved classes of mRNAs that are enriched on microtubules in both human and X. laevis. Active mitotic translation occurs on X. laevis meiotic spindles, and a subset of microtubule-bound mRNAs (MT-mRNAs) associate with polyribosomes. Although many MT-mRNAs associate with polyribosomes, we find that active translation is not required for mRNA localization to mitotic microtubules. Our results represent the first genome-wide survey of mRNAs localized to a specific cytoskeletal component and suggest that microtubule localization of specific mRNAs is likely to function in mitotic regulation and mRNA segregation during cell division.
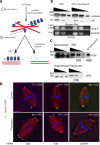
Figures
References
-
- Blower, M.D., M. Nachury, R. Heald, and K. Weis. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 121:223–234. - PubMed
-
- Desai, A., A. Murray, T.J. Mitchison, and C.E. Walczak. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. - PubMed

