Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition
- PMID: 18166650
- PMCID: PMC2373499
- DOI: 10.1083/jcb.200705160
Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition
Abstract
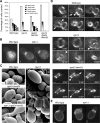
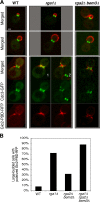
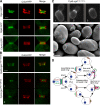
Cells of the budding yeast Saccharomyces cerevisiae are born carrying localized transmembrane landmark proteins that guide the subsequent establishment of a polarity axis and hence polarized growth to form a bud in the next cell cycle. In haploid cells, the relevant landmark proteins are concentrated at the site of the preceding cell division, to which they recruit Cdc24, the guanine nucleotide exchange factor for the conserved polarity regulator Cdc42. However, instead of polarizing at the division site, the new polarity axis is directed next to but not overlapping that site. Here, we show that the Cdc42 guanosine triphosphatase-activating protein (GAP) Rga1 establishes an exclusion zone at the division site that blocks subsequent polarization within that site. In the absence of localized Rga1 GAP activity, new buds do in fact form within the old division site. Thus, Cdc42 activators and GAPs establish concentric zones of action such that polarization is directed to occur adjacent to but not within the previous cell division site.
Figures

References
-
- Ahmadian, M.R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4:686–689. - PubMed
-
- Barton, A.A. 1950. Some aspects of cell division in Saccharomyces cerevisiae. J. Gen. Microbiol. 4:84–86. - PubMed
-
- Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 1603:47–82. - PubMed
-
- Bernards, A., and J. Settleman. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14:377–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

