Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages
- PMID: 18167562
- PMCID: PMC2151136
- DOI: 10.1371/journal.pone.0001403
Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages
Abstract
Background: Transcriptional profiling using microarrays provides a unique opportunity to decipher host pathogen cross-talk on the global level. Here, for the first time, we have been able to investigate gene expression changes in both Mycobacterium tuberculosis, a major human pathogen, and its human host cells, macrophages and dendritic cells.
Methodology/principal findings: In addition to common responses, we could identify eukaryotic and microbial transcriptional signatures that are specific to the cell type involved in the infection process. In particular M. tuberculosis shows a marked stress response when inside dendritic cells, which is in accordance with the low permissivity of these specialized phagocytes to the tubercle bacillus and to other pathogens. In contrast, the mycobacterial transcriptome inside macrophages reflects that of replicating bacteria. On the host cell side, differential responses to infection in macrophages and dendritic cells were identified in genes involved in oxidative stress, intracellular vesicle trafficking and phagosome acidification.
Conclusions/significance: This study provides the proof of principle that probing the host and the microbe transcriptomes simultaneously is a valuable means to accessing unique information on host pathogen interactions. Our results also underline the extraordinary plasticity of host cell and pathogen responses to infection, and provide a solid framework to further understand the complex mechanisms involved in immunity to M. tuberculosis and in mycobacterial adaptation to different intracellular environments.
Conflict of interest statement
Figures
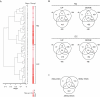
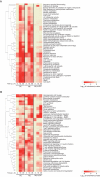


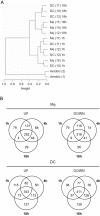
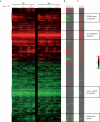

References
-
- Ricciardi-Castagnoli P, Granucci F. Opinion: Interpretation of the complexity of innate immune responses by functional genomics. Nat Rev Immunol. 2002;2:881–889. - PubMed
-
- Schnappinger D, Schoolnik GK, Ehrt S. Expression profiling of host pathogen interactions: how Mycobacterium tuberculosis and the macrophage adapt to one another. Microbes Infect. 2006;8:1132–1140. - PubMed
-
- Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–577. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

