A simple principle concerning the robustness of protein complex activity to changes in gene expression
- PMID: 18171472
- PMCID: PMC2242779
- DOI: 10.1186/1752-0509-2-1
A simple principle concerning the robustness of protein complex activity to changes in gene expression
Abstract
Background: The functions of a eukaryotic cell are largely performed by multi-subunit protein complexes that act as molecular machines or information processing modules in cellular networks. An important problem in systems biology is to understand how, in general, these molecular machines respond to perturbations.
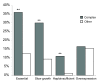
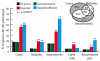
Results: In yeast, genes that inhibit growth when their expression is reduced are strongly enriched amongst the subunits of multi-subunit protein complexes. This applies to both the core and peripheral subunits of protein complexes, and the subunits of each complex normally have the same loss-of-function phenotypes. In contrast, genes that inhibit growth when their expression is increased are not enriched amongst the core or peripheral subunits of protein complexes, and the behaviour of one subunit of a complex is not predictive for the other subunits with respect to over-expression phenotypes.
Conclusion: We propose the principle that the overall activity of a protein complex is in general robust to an increase, but not to a decrease in the expression of its subunits. This means that whereas phenotypes resulting from a decrease in gene expression can be predicted because they cluster on networks of protein complexes, over-expression phenotypes cannot be predicted in this way. We discuss the implications of these findings for understanding how cells are regulated, how they evolve, and how genetic perturbations connect to disease in humans.
Figures

Comment in
-
On gene dosage balance in protein complexes: a comment on Semple JI, Vavouri T, Lehner B. A simple principle concerning the robustness of protein complex activity to changes in gene expression.BMC Syst Biol. 2009 Jan 30;3:16. doi: 10.1186/1752-0509-3-16. BMC Syst Biol. 2009. PMID: 19183469 Free PMC article.
References
-
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. - DOI - PubMed
-
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. - DOI - PubMed
-
- Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, Li N, Mani R, Hyman AA, Sonnichsen B, Echeverri CJ, Roth FP, Vidal M, Piano F. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

