High-resolution analysis of condition-specific regulatory modules in Saccharomyces cerevisiae
- PMID: 18171483
- PMCID: PMC2395236
- DOI: 10.1186/gb-2008-9-1-r2
High-resolution analysis of condition-specific regulatory modules in Saccharomyces cerevisiae
Abstract
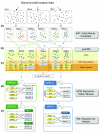
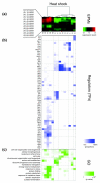
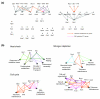
We present an approach for identifying condition-specific regulatory modules by using separate units of gene expression profiles along with ChIP-chip and motif data from Saccharomyces cerevisiae. By investigating the unique and common features of the obtained condition-specific modules, we detected several important properties of transcriptional network reorganization. Our approach reveals the functionally distinct coregulated submodules embedded in a coexpressed gene module and provides an effective method for identifying various condition-specific regulatory events at high resolution.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

