Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'
- PMID: 18172165
- PMCID: PMC2151014
- DOI: 10.1101/gad.1622708
Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'
Abstract
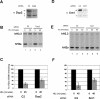
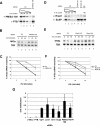
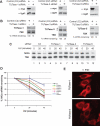
Histone mRNAs are rapidly degraded at the end of S phase or when DNA replication is inhibited. Histone mRNAs end in a conserved stem-loop rather than a poly(A) tail. Degradation of histone mRNAs requires the stem-loop sequence, which binds the stem-loop-binding protein (SLBP), active translation of the histone mRNA, and the location of the stem-loop close to the termination codon. We report that the initial step in histone mRNA degradation is the addition of uridines to the 3' end of the histone mRNA, both after inhibition of DNA replication and at the end of S phase. Lsm1 is required for histone mRNA degradation and is present in a complex containing SLBP on the 3' end of histone mRNA after inhibition of DNA replication. We cloned degradation intermediates that had been partially degraded from both the 5' and the 3' ends. RNAi experiments demonstrate that both the exosome and 5'-to-3' decay pathway components are required for degradation, and individual histone mRNAs are then degraded simultaneously 5' to 3' and 3' to 5'.
Figures




Comment in
-
New ways to meet your (3') end oligouridylation as a step on the path to destruction.Genes Dev. 2008 Jan 1;22(1):1-7. doi: 10.1101/gad.1634508. Genes Dev. 2008. PMID: 18172159 Free PMC article. Review. No abstract available.
References
-
- Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. - PubMed
-
- Blum E., Carpousis A.J., Higgins C.F. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 1999;274:4009–4016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
