Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson's disease
- PMID: 18173235
- PMCID: PMC3319057
- DOI: 10.1021/pr070546l
Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson's disease
Abstract
The molecular mechanisms underlying the changes in the nigrostriatal pathway in Parkinson's disease (PD) are not completely understood. Here, we use mass spectrometry and microarrays to study the proteomic and transcriptomic changes in the striatum of two mouse models of PD, induced by the distinct neurotoxins 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and methamphetamine (METH). Proteomic analyses resulted in the identification and relative quantification of 912 proteins with two or more unique peptides and 86 proteins with significant abundance changes following neurotoxin treatment. Similarly, microarray analyses revealed 181 genes with significant changes in mRNA, following neurotoxin treatment. The combined protein and gene list provides a clearer picture of the potential mechanisms underlying neurodegeneration observed in PD. Functional analysis of this combined list revealed a number of significant categories, including mitochondrial dysfunction, oxidative stress response, and apoptosis. These results constitute one of the largest descriptive data sets integrating protein and transcript changes for these neurotoxin models with many similar end point phenotypes but distinct mechanisms.
Figures

 0.91 and 0.89, respectively). (B) Heatmap of proteins identified as significantly differentially regulated (p < 0.05). The regulated proteins show consistency across samples. (C) Multiple peptides detected from individual proteins show good consistency in expression levels.
0.91 and 0.89, respectively). (B) Heatmap of proteins identified as significantly differentially regulated (p < 0.05). The regulated proteins show consistency across samples. (C) Multiple peptides detected from individual proteins show good consistency in expression levels.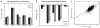
 0.6563) is significant (p < 10−20).
0.6563) is significant (p < 10−20).
 0.718, p < 10−16). (D) Selected gene expression levels confirmed using quantitative real-time polymerase chain reaction (qRT-PCR) (p < 0.05, all tests). (E) mRNA levels compared to protein abundance. A scatter plot comparing protein to mRNA levels averaged across treated and control samples. LOWESS fitted curve (dark line) (R
0.718, p < 10−16). (D) Selected gene expression levels confirmed using quantitative real-time polymerase chain reaction (qRT-PCR) (p < 0.05, all tests). (E) mRNA levels compared to protein abundance. A scatter plot comparing protein to mRNA levels averaged across treated and control samples. LOWESS fitted curve (dark line) (R
 0.289, p < 10−21). Efficiently transcribed genes lie > 2 standard deviations (SD) above the LOWESS curve (upper gray line indicates 2 SD cutoff), and inefficiently transcribed genes lie < 1.4 SD below the curve (lower gray line).
0.289, p < 10−21). Efficiently transcribed genes lie > 2 standard deviations (SD) above the LOWESS curve (upper gray line indicates 2 SD cutoff), and inefficiently transcribed genes lie < 1.4 SD below the curve (lower gray line).References
-
- Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci. 1973;20(4):415–455. - PubMed
-
- Marsden CD. Parkinson’s disease. Lancet. 1990;335(8695):948–952. - PubMed
-
- Gandhi S, Wood NW. Molecular pathogenesis of Parkinson’s disease. Hum Mol Genet. 2005;14(18):2749–2755. - PubMed
-
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. - PubMed
-
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48(1):77–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

