Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres
- PMID: 18174443
- PMCID: PMC2586718
- DOI: 10.1126/science.1150944
Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres
Abstract
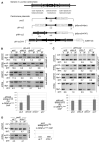
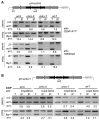
Heterochromatin is defined by distinct posttranslational modifications on histones, such as methylation of histone H3 at lysine 9 (H3K9), which allows heterochromatin protein 1 (HP1)-related chromodomain proteins to bind. Heterochromatin is frequently found near CENP-A chromatin, which is the key determinant of kinetochore assembly. We have discovered that the RNA interference (RNAi)-directed heterochromatin flanking the central kinetochore domain at fission yeast centromeres is required to promote CENP-A(Cnp1) and kinetochore assembly over the central domain. The H3K9 methyltransferase Clr4 (Suv39); the ribonuclease Dicer, which cleaves heterochromatic double-stranded RNA to small interfering RNA (siRNA); Chp1, a component of the RNAi effector complex (RNA-induced initiation of transcriptional gene silencing; RITS); and Swi6 (HP1) are required to establish CENP-A(Cnp1) chromatin on naïve templates. Once assembled, CENP-A(Cnp1) chromatin is propagated by epigenetic means in the absence of heterochromatin. Thus, another, potentially conserved, role for centromeric RNAi-directed heterochromatin has been identified.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

