The NR3B subgroup: an ovERRview
- PMID: 18174917
- PMCID: PMC2121319
- DOI: 10.1621/nrs.05009
The NR3B subgroup: an ovERRview
Abstract
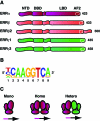
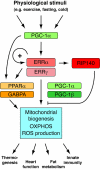
Members of the NR3B group of the nuclear receptor superfamily, known as the estrogen-related receptors (ERRs), were the first orphan receptors to be identified two decades ago. Despite the fact that a natural ligand has yet to be associated with the ERRs, considerable knowledge about their mode of action and biological functions has emerged through extensive biochemical, genetic and functional genomics studies. This review describes our current understanding of how the ERRs work as transcription factors and as such, how they control diverse developmental and physiological programs.
Figures
References
-
- Alaynick W. A., Kondo R. P., Xie W., He W., Dufour C. R., Downes M., Jonker J. W., Giles W., Naviaux R. K., Giguere V., Evans R. M. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. - PubMed
-
- Araki M., Motojima K. Identification of ERRalpha as a specific partner of PGC-1alpha for the activation of PDK4 gene expression in muscle. Febs J. 2006;273:1669–80. - PubMed
-
- Ariazi E. A., Clark G. M., Mertz J. E. Estrogen-related receptor α and estrogen-related receptor γ associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–8. - PubMed
-
- Ariazi E. A., Kraus R. J., Farrell M. L., Jordan V. C., Mertz J. E. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res. 2007;5:71–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

