A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex
- PMID: 18176557
- PMCID: PMC3065498
- DOI: 10.1038/nchembio.63
A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex
Abstract
The MRN (Mre11-Rad50-Nbs1)-ATM (ataxia-telangiectasia mutated) pathway is essential for sensing and signaling from DNA double-strand breaks. The MRN complex acts as a DNA damage sensor, maintains genome stability during DNA replication, promotes homology-dependent DNA repair and activates ATM. MRN is essential for cell viability, which has limited functional studies of the complex. Small-molecule inhibitors of MRN could circumvent this experimental limitation and could also be used as cellular radio- and chemosensitization compounds. Using cell-free systems that recapitulate faithfully the MRN-ATM signaling pathway, we designed a forward chemical genetic screen to identify inhibitors of the pathway, and we isolated 6-(4-hydroxyphenyl)-2-thioxo-2,3-dihydro-4(1H)-pyrimidinone (mirin, 1) as an inhibitor of MRN. Mirin prevents MRN-dependent activation of ATM without affecting ATM protein kinase activity, and it inhibits Mre11-associated exonuclease activity. Consistent with its ability to target the MRN complex, mirin abolishes the G2/M checkpoint and homology-dependent repair in mammalian cells.
Figures

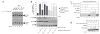
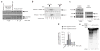
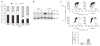

Comment in
-
Small molecule versus DNA repair nanomachine.Nat Chem Biol. 2008 Feb;4(2):86-8. doi: 10.1038/nchembio0208-86. Nat Chem Biol. 2008. PMID: 18202674 Free PMC article.
-
Corrected structure of mirin, a small-molecule inhibitor of the Mre11-Rad50-Nbs1 complex.Nat Chem Biol. 2009 Mar;5(3):129-30; author reply 130. doi: 10.1038/nchembio0309-129. Nat Chem Biol. 2009. PMID: 19219009 Free PMC article. No abstract available.
References
-
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. - PubMed
-
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. - PubMed
-
- Lavin MF, et al. ATM signaling and genomic stability in response to DNA damage. Mutat Res. 2005;569:123–132. - PubMed
-
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

