Interaction of transmembrane helices in ATP synthase subunit a in solution as revealed by spin label difference NMR
- PMID: 18178144
- PMCID: PMC2276983
- DOI: 10.1016/j.bbabio.2007.11.011
Interaction of transmembrane helices in ATP synthase subunit a in solution as revealed by spin label difference NMR
Abstract
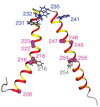
Subunit a in the membrane traversing F0 sector of Escherichia coli ATP synthase is known to fold with five transmembrane helices (TMHs) with residue 218 in TMH IV packing close to residue 248 in TMH V. In this study, we have introduced a spin label probe at Cys residues substituted at positions 222 or 223 and measured the effects on the Trp epsilon NH indole NMR signals of the seven Trp residues in the protein. The protein was purified and NMR experiments were carried out in a chloroform-methanol-H2O (4:4:1) solvent mixture. The spin label at positions 222 or 223 proved to broaden the signals of W231, W232, W235 and W241 located at the periplasmic ends of TMH IV and TMH V and the connecting loop between these helices. The broadening of W241 would require that the loop residues fold back on themselves in a hairpin-like structure much like it is predicted to fold in the native membrane. Placement of the spin label probe at several other positions also proved to have broadening effects on some of these Trp residues and provided additional constraints on folding of TMH IV and TMH V. The effects of the 223 probes on backbone amide resonances of subunit a were also measured by an HNCO experiment and the results are consistent with the two helices folding back on themselves in this solvent mixture. When Cys and Trp were substituted at residues 206 and 254 at the cytoplasmic ends of TMHs IV and V respectively, the W254 resonance was not broadened by the spin label at position 206. We conclude that the helices fold back on themselves in this solvent system and then pack at an angle such that the cytoplasmic ends of the polypeptide backbone are significantly displaced from each other.
Figures








Similar articles
-
Structure of Ala24/Asp61 --> Asp24/Asn61 substituted subunit c of Escherichia coli ATP synthase: implications for the mechanism of proton transport and rotary movement in the F0 complex.Biochemistry. 2002 Apr 30;41(17):5537-47. doi: 10.1021/bi012198l. Biochemistry. 2002. PMID: 11969414
-
Folded state of the integral membrane colicin E1 immunity protein in solvents of mixed polarity.Biochemistry. 2000 Oct 10;39(40):12131-9. doi: 10.1021/bi000206c. Biochemistry. 2000. PMID: 11015191
-
Structure of Ala(20) --> Pro/Pro(64) --> Ala substituted subunit c of Escherichia coli ATP synthase in which the essential proline is switched between transmembrane helices.J Biol Chem. 2001 Jul 20;276(29):27449-54. doi: 10.1074/jbc.M100762200. Epub 2001 Apr 30. J Biol Chem. 2001. PMID: 11331283
-
Structural model of the transmembrane Fo rotary sector of H+-transporting ATP synthase derived by solution NMR and intersubunit cross-linking in situ.Biochim Biophys Acta. 2002 Oct 11;1565(2):232-45. doi: 10.1016/s0005-2736(02)00572-2. Biochim Biophys Acta. 2002. PMID: 12409198 Review.
-
Subunit organization and structure in the F0 sector of Escherichia coli F1F0 ATP synthase.Biochim Biophys Acta. 1998 Jun 10;1365(1-2):135-42. doi: 10.1016/s0005-2728(98)00053-x. Biochim Biophys Acta. 1998. PMID: 9693732 Review.
Cited by
-
Interaction with monomeric subunit c drives insertion of ATP synthase subunit a into the membrane and primes a-c complex formation.J Biol Chem. 2011 Nov 4;286(44):38583-38591. doi: 10.1074/jbc.M111.294868. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21900248 Free PMC article.
-
The proton-translocating a subunit of F0F1-ATP synthase is allocated asymmetrically to the peripheral stalk.J Biol Chem. 2008 Nov 28;283(48):33602-10. doi: 10.1074/jbc.M805170200. Epub 2008 Sep 11. J Biol Chem. 2008. PMID: 18786919 Free PMC article.
-
NMR and mutational identification of the collagen-binding site of the chaperone Hsp47.PLoS One. 2012;7(9):e45930. doi: 10.1371/journal.pone.0045930. Epub 2012 Sep 25. PLoS One. 2012. PMID: 23049894 Free PMC article.
-
Resolving the negative potential side (n-side) water-accessible proton pathway of F-type ATP synthase by molecular dynamics simulations.J Biol Chem. 2012 Oct 19;287(43):36536-43. doi: 10.1074/jbc.M112.398396. Epub 2012 Aug 31. J Biol Chem. 2012. PMID: 22942277 Free PMC article.
References
-
- Yoshida M, Muneyuki E, Hisabori T. ATP synthase - a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. - PubMed
-
- Capaldi RA, Aggeler, R R. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 2002;27:154–160. - PubMed
-
- Senior AE. ATP synthesis by oxidative phosphorylation. Physiol. Rev. 1988;68:177–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

