Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution
- PMID: 18178768
- PMCID: PMC2254919
- DOI: 10.1105/tpc.107.056309
Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution
Abstract
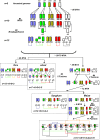
The grass family comprises the most important cereal crops and is a good system for studying, with comparative genomics, mechanisms of evolution, speciation, and domestication. Here, we identified and characterized the evolution of shared duplications in the rice (Oryza sativa) and wheat (Triticum aestivum) genomes by comparing 42,654 rice gene sequences with 6426 mapped wheat ESTs using improved sequence alignment criteria and statistical analysis. Intraspecific comparisons identified 29 interchromosomal duplications covering 72% of the rice genome and 10 duplication blocks covering 67.5% of the wheat genome. Using the same methodology, we assessed orthologous relationships between the two genomes and detected 13 blocks of colinearity that represent 83.1 and 90.4% of the rice and wheat genomes, respectively. Integration of the intraspecific duplications data with colinearity relationships revealed seven duplicated segments conserved at orthologous positions. A detailed analysis of the length, composition, and divergence time of these duplications and comparisons with sorghum (Sorghum bicolor) and maize (Zea mays) indicated common and lineage-specific patterns of conservation between the different genomes. This allowed us to propose a model in which the grass genomes have evolved from a common ancestor with a basic number of five chromosomes through a series of whole genome and segmental duplications, chromosome fusions, and translocations.
Figures

References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Bennetzen, J.L. (2005). Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15 621–627. - PubMed
-
- Blake, N.K., Lehfeldt, B.R., Lavin, M., and Talbert, L.E. (1999). Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: The B genome of wheat. Genome 42 351–360. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

