Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response
- PMID: 18178771
- PMCID: PMC2254933
- DOI: 10.1105/tpc.107.051862
Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response
Abstract
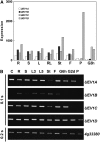
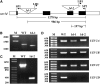
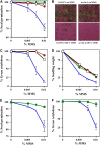
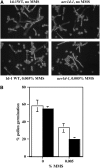
DNA damage tolerance (DDT) in budding yeast requires Lys-63-linked polyubiquitination of the proliferating cell nuclear antigen. The ubiquitin-conjugating enzyme Ubc13 and the Ubc enzyme variant (Uev) methyl methanesulfonate2 (Mms2) are required for this process. Mms2 homologs have been found in all eukaryotic genomes examined; however, their roles in multicellular eukaryotes have not been elucidated. We report the isolation and characterization of four UEV1 genes from Arabidopsis thaliana. All four Uev1 proteins can form a stable complex with At Ubc13 or with Ubc13 from yeast or human and can promote Ubc13-mediated Lys-63 polyubiquitination. All four Uev1 proteins can replace yeast MMS2 DDT functions in vivo. Although these genes are ubiquitously expressed in most tissues, UEV1D appears to express at a much higher level in germinating seeds and in pollen. We obtained and characterized two uev1d null mutant T-DNA insertion lines. Compared with wild-type plants, seeds from uev1d null plants germinated poorly when treated with a DNA-damaging agent. Those that germinated grew slower, and the majority ceased growth within 2 weeks. Pollen from uev1d plants also displayed a moderate but significant decrease in germination in the presence of DNA damage. This report links Ubc13-Uev with functions in DNA damage response in Arabidopsis.
Figures



References
-
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6 463–470. - PubMed
-
- Barbour, L., and Xiao, W. (2003). Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: A yeast model. Mutat. Res. 532 137–155. - PubMed
-
- Bartel, P.L., and Fields, S. (1995). Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254 241–263. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

