An open-source representation for 2-DE-centric proteomics and support infrastructure for data storage and analysis
- PMID: 18179696
- PMCID: PMC2231339
- DOI: 10.1186/1471-2105-9-4
An open-source representation for 2-DE-centric proteomics and support infrastructure for data storage and analysis
Abstract
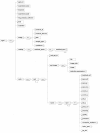
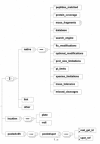
Background: In spite of two-dimensional gel electrophoresis (2-DE) being an effective and widely used method to screen the proteome, its data standardization has still not matured to the level of microarray genomics data or mass spectrometry approaches. The trend toward identifying encompassing data standards has been expanding from genomics to transcriptomics, and more recently to proteomics. The relative success of genomic and transcriptomic data standardization has enabled the development of central repositories such as GenBank and Gene Expression Omnibus. An equivalent 2-DE-centric data structure would similarly have to include a balance among raw data, basic feature detection results, sufficiency in the description of the experimental context and methods, and an overall structure that facilitates a diversity of usages, from central reposition to local data representation in LIMs systems.
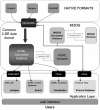
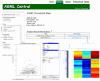
Results & conclusion: Achieving such a balance can only be accomplished through several iterations involving bioinformaticians, bench molecular biologists, and the manufacturers of the equipment and commercial software from which the data is primarily generated. Such an encompassing data structure is described here, developed as the mature successor to the well established and broadly used earlier version. A public repository, AGML Central, is configured with a suite of tools for the conversion from a variety of popular formats, web-based visualization, and interoperation with other tools and repositories, and is particularly mass-spectrometry oriented with I/O for annotation and data analysis.
Figures
References
-
- Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. - DOI - PubMed
-
- mzData http://www.psidev.info/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

