A statistical method for predicting classical HLA alleles from SNP data
- PMID: 18179884
- PMCID: PMC2253983
- DOI: 10.1016/j.ajhg.2007.09.001
A statistical method for predicting classical HLA alleles from SNP data
Abstract
Genetic variation at classical HLA alleles is a crucial determinant of transplant success and susceptibility to a large number of infectious and autoimmune diseases. However, large-scale studies involving classical type I and type II HLA alleles might be limited by the cost of allele-typing technologies. Although recent studies have shown that some common HLA alleles can be tagged with small numbers of markers, SNP-based tagging does not offer a complete solution to predicting HLA alleles. We have developed a new statistical methodology to use SNP variation within the region to predict alleles at key class I (HLA-A, HLA-B, and HLA-C) and class II (HLA-DRB1, HLA-DQA1, and HLA-DQB1) loci. Our results indicate that a single panel of approximately 100 SNPs typed across the region is sufficient for predicting both rare and common HLA alleles with up to 95% accuracy in both African and non-African populations. Furthermore, we show that HLA alleles can be successfully predicted by using previously genotyped SNPs that are within the MHC and that had not been chosen for their ability to predict HLA alleles, such as those included on genome-wide products. These results indicate that our methodology, combined with an extended database of reference haplotypes, will facilitate large-scale experiments, including disease-association studies and vaccine trials, in which detailed information about HLA type is valuable.
Figures

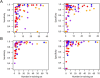
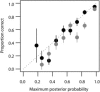
References
-
- Malkki M., Single R., Carrington M., Thomson G., Petersdorf E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: Implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 2005;66:114–124. - PubMed
-
- Cooke G.S., Hill A.V. Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2001;2:967–977. - PubMed
-
- Gregersen P.K., Behrens T.W. Genetics of autoimmune diseases–disorders of immune homeostasis. Nat. Rev. Genet. 2006;7:917–928. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

