An X-linked myopathy with postural muscle atrophy and generalized hypertrophy, termed XMPMA, is caused by mutations in FHL1
- PMID: 18179888
- PMCID: PMC2253986
- DOI: 10.1016/j.ajhg.2007.09.004
An X-linked myopathy with postural muscle atrophy and generalized hypertrophy, termed XMPMA, is caused by mutations in FHL1
Abstract
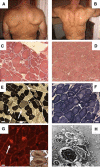
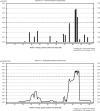
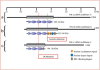
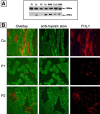
We have identified a large multigenerational Austrian family displaying a novel form of X-linked recessive myopathy. Affected individuals develop an adult-onset scapulo-axio-peroneal myopathy with bent-spine syndrome characterized by specific atrophy of postural muscles along with pseudoathleticism or hypertrophy and cardiac involvement. Known X-linked myopathies were excluded by simple-tandem-repeat polymorphism (STRP) and single-nucleotide polymorphism (SNP) analysis, direct gene sequencing, and immunohistochemical analysis. STRP analysis revealed significant linkage at Xq25-q27.1. Haplotype analysis based on SNP microarray data from selected family members confirmed this linkage region on the distal arm of the X chromosome, thereby narrowing down the critical interval to 12 Mb. Sequencing of functional candidate genes led to the identification of a missense mutation within the four and a half LIM domain 1 gene (FHL1), which putatively disrupts the fourth LIM domain of the protein. Mutation screening of FHL1 in a myopathy family from the UK exhibiting an almost identical phenotype revealed a 3 bp insertion mutation within the second LIM domain. FHL1 on Xq26.3 is highly expressed in skeletal and cardiac muscles. Western-blot analysis of muscle biopsies showed a marked decrease in protein expression of FHL1 in patients, in concordance with the genetic data. In summary, we have to our knowledge characterized a new disorder, X-linked myopathy with postural muscle atrophy (XMPMA), and identified FHL1 as the causative gene. This is the first FHL protein to be identified in conjunction with a human genetic disorder and further supports the role of FHL proteins in the development and maintenance of muscle tissue. Mutation screening of FHL1 should be considered for patients with uncharacterized myopathies and cardiomyopathies.
Figures



References
-
- Davies K.E., Nowak K.J. Molecular mechanisms of muscular dystrophies: Old and new players. Nat. Rev. Mol. Cell Biol. 2006;7:762–773. - PubMed
-
- Kalimo H., Savontaus M.-L., Lang H., Paljarvi L., Sonninen V., Dean P.B., Katevuo K., Salminen A. X-linked myopathy with excessive autophagy: A new hereditary muscle disease. Ann. Neurol. 1988;23:258–265. - PubMed
-
- Danek A., Rubio J.P., Rampoldi L., Ho M., Dobson-Stone C., Tison F., Symmans W.A., Oechsner M., Kalckreuth W., Watt J.M. McLeod neuroacanthocytosis: Genotype and phenotype. Ann. Neurol. 2001;50:755–764. - PubMed
-
- Schwarzmeier J., Moser K., Lujf A. Pyruvate kinase deficiency of erythrocytes in a case of hereditary myopathy. Klin. Wochenschr. 1971;49:156–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

