Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability
- PMID: 18180284
- PMCID: PMC2258774
- DOI: 10.1128/MCB.01717-07
Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability
Abstract
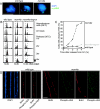
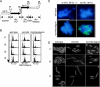
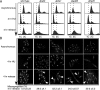
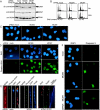
The minichromosome maintenance (MCM) complex plays essential, conserved roles throughout DNA synthesis: first, as a component of the prereplication complex at origins and, then, as a helicase associated with replication forks. Here we use fission yeast (Schizosaccharomyces pombe) as a model to demonstrate a role for the MCM complex in protecting replication fork structure and promoting recovery from replication arrest. Loss of MCM function generates lethal double-strand breaks at sites of DNA synthesis during replication elongation, suggesting replication fork collapse. MCM function also maintains the stability of forks stalled by hydroxyurea that activate the replication checkpoint. In cells where the checkpoint is activated, Mcm4 binds the Cds1 kinase and undergoes Cds1-dependent phosphorylation. MCM proteins also interact with proteins involved in homologous recombination, which promotes recovery from arrest by ensuring normal mitosis. We suggest that the MCM complex links replication fork stabilization with checkpoint arrest and recovery through direct interactions with checkpoint and recombination proteins and that this role in S-phase genome stability is conserved from yeast to human cells.
Figures


References
-
- Bailis, J. M., P. Bernard, R. Antonelli, R. Allshire, and S. L. Forsburg. 2003. Hsk1/Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 1111-1116. - PubMed
-
- Bailis, J. M., and S. L. Forsburg. 2004. MCM proteins: DNA damage, mutagenesis, repair. Curr. Opin. Gen. Dev. 1417-21. - PubMed
-
- Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434864-870. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
