Signaling networks assembled by oncogenic EGFR and c-Met
- PMID: 18180459
- PMCID: PMC2206598
- DOI: 10.1073/pnas.0707270105
Signaling networks assembled by oncogenic EGFR and c-Met
Abstract
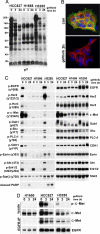
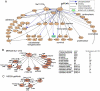
A major question regarding the sensitivity of solid tumors to targeted kinase inhibitors is why some tumors respond and others do not. The observation that many tumors express EGF receptor (EGFR), yet only a small subset with EGFR-activating mutations respond clinically to EGFR inhibitors (EGFRIs), suggests that responsive tumors uniquely depend on EGFR signaling for their survival. The nature of this dependence is not understood. Here, we investigate dependence on EGFR signaling by comparing non-small-cell lung cancer cell lines driven by EGFR-activating mutations and genomic amplifications using a global proteomic analysis of phospho-tyrosine signaling. We identify an extensive receptor tyrosine kinase signaling network established in cells expressing mutated and activated EGFR or expressing amplified c-Met. We show that in drug sensitive cells the targeted tyrosine kinase drives other RTKs and an extensive network of downstream signaling that collapse with drug treatment. Comparison of the signaling networks in EGFR and c-Met-dependent cells identify a "core network" of approximately 50 proteins that participate in pathways mediating drug response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
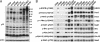
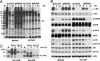
References
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. Science. 2004;304:1497–1500. - PubMed
-
- Riely GJ, Politi KA, Miller VA, Pao W. Clin Cancer Res. 2006;12:7232–7241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

