Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution
- PMID: 18182495
- PMCID: PMC2206580
- DOI: 10.1073/pnas.0707318105
Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution
Abstract
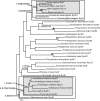
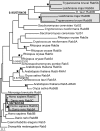
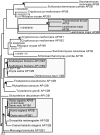
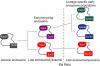
The process by which some eukaryotic organelles, for example the endomembrane system, evolved without endosymbiotic input remains poorly understood. This problem largely arises because many major cellular systems predate the last common eukaryotic ancestor (LCEA) and thus do not provide examples of organellogenesis in progress. A model is emerging whereby gene duplication and divergence of multiple "specificity-" or "identity-" encoding proteins for the various endomembranous organelles produced the diversity of nonendosymbiotically derived cellular compartments present in modern eukaryotes. To address this possibility, we analyzed three molecular components of the endocytic membrane-trafficking machinery. Phylogenetic analyses of the endocytic syntaxins, Rab 5, and the beta-adaptins each reveal a pattern of ancestral, undifferentiated endocytic homologues in the LCEA. Subsequently, these undifferentiated progenitors independently duplicated in widely divergent lineages, convergently producing components with similar endocytic roles, e.g., beta1 and beta2-adaptin. In contrast, beta3, beta4, and all other adaptin complex subunits, as well as paralogues of the syntaxins and Rabs specific for the other membrane-trafficking organelles, all evolved before the LCEA. Thus, the process giving rise to the differentiated organelles of the endocytic system appears to have been interrupted by the major speciation event that produced the extant eukaryotic lineages. These results suggest that although many endocytic components evolved before the LCEA, other major features evolved independently and convergently after diversification into the primary eukaryotic supergroups. This finding provides an example of a basic cellular system that was simpler in the LCEA than in many extant eukaryotes and yields insight into nonendosymbiotic organelle evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Butterfield NJ. Paleobiology. 2000;26:386–404.
-
- Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. New Haven, CT: Yale Univ Press; 1970.
-
- Cavalier-Smith T. Curr Opin Microbiol. 2002;5:612–619. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

