A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis
- PMID: 18183306
- PMCID: PMC2173943
- DOI: 10.1371/journal.pone.0001426
A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis
Abstract
Background: Diatoms are unicellular algae responsible for approximately 20% of global carbon fixation. Their evolution by secondary endocytobiosis resulted in a complex cellular structure and metabolism compared to algae with primary plastids.
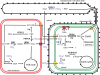
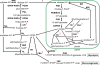
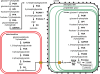
Methodology/principal findings: The whole genome sequence of the diatom Phaeodactylum tricornutum has recently been completed. We identified and annotated genes for enzymes involved in carbohydrate pathways based on extensive EST support and comparison to the whole genome sequence of a second diatom, Thalassiosira pseudonana. Protein localization to mitochondria was predicted based on identified similarities to mitochondrial localization motifs in other eukaryotes, whereas protein localization to plastids was based on the presence of signal peptide motifs in combination with plastid localization motifs previously shown to be required in diatoms. We identified genes potentially involved in a C4-like photosynthesis in P. tricornutum and, on the basis of sequence-based putative localization of relevant proteins, discuss possible differences in carbon concentrating mechanisms and CO(2) fixation between the two diatoms. We also identified genes encoding enzymes involved in photorespiration with one interesting exception: glycerate kinase was not found in either P. tricornutum or T. pseudonana. Various Calvin cycle enzymes were found in up to five different isoforms, distributed between plastids, mitochondria and the cytosol. Diatoms store energy either as lipids or as chrysolaminaran (a beta-1,3-glucan) outside of the plastids. We identified various beta-glucanases and large membrane-bound glucan synthases. Interestingly most of the glucanases appear to contain C-terminal anchor domains that may attach the enzymes to membranes.
Conclusions/significance: Here we present a detailed synthesis of carbohydrate metabolism in diatoms based on the genome sequences of Thalassiosira pseudonana and Phaeodactylum tricornutum. This model provides novel insights into acquisition of dissolved inorganic carbon and primary metabolic pathways of carbon in two different diatoms, which is of significance for an improved understanding of global carbon cycles.
Conflict of interest statement
Figures

References
-
- Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–205. - PubMed
-
- Brzezinski MA, Pride CJ, Franck VM. A switch from Si(OH)4 to NO3-depletion in the glacial Southern Ocean. Geophys. Res Let. 2002;29:5-1 to 5-5.
-
- Granum E, Raven JA, Leegood RC. How do marine diatoms fix 10 billion tonnes of anorganic carbon per year. Can J Bot. 2005;83:898–908.
-
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, et al. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot. 1998;76:1052–1071.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

