Envelope protein palmitoylations are crucial for murine coronavirus assembly
- PMID: 18184706
- PMCID: PMC2258982
- DOI: 10.1128/JVI.01906-07
Envelope protein palmitoylations are crucial for murine coronavirus assembly
Abstract
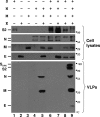
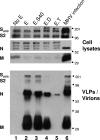
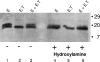
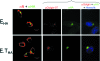
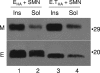
The coronavirus assembly process encloses a ribonucleoprotein genome into vesicles containing the lipid-embedded proteins S (spike), E (envelope), and M (membrane). This process depends on interactions with membranes that may involve palmitoylation, a common posttranslational lipidation of cysteine residues. To determine whether specific palmitoylations influence coronavirus assembly, we introduced plasmid DNAs encoding mouse hepatitis coronavirus (MHV) S, E, M, and N (nucleocapsid) into 293T cells and found that virus-like particles (VLPs) were robustly assembled and secreted into culture medium. Palmitate adducts predicted on cysteines 40, 44, and 47 of the 83-residue E protein were then evaluated by constructing mutant cDNAs with alanine or glycine codon substitutions at one or more of these positions. Triple-substituted proteins (E.Ts) lacked palmitate adducts. Both native E and E.T proteins localized at identical perinuclear locations, and both copurified with M proteins, but E.T was entirely incompetent for VLP production. In the presence of the E.T proteins, the M protein subunits accumulated into detergent-insoluble complexes that failed to secrete from cells, while native E proteins mobilized M into detergent-soluble secreted forms. Many of these observations were corroborated in the context of natural MHV infections, with native E, but not E.T, complementing debilitated recombinant MHVs lacking E. Our findings suggest that palmitoylations are essential for E to act as a vesicle morphogenetic protein and further argue that palmitoylated E proteins operate by allowing the primary coronavirus assembly subunits to assume configurations that can mobilize into secreted lipid vesicles and virions.
Figures



References
-
- Bauer, M., and L. Pelkmans. 2006. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 5805559-5564. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

