Preferred side-chain constellations at antiparallel coiled-coil interfaces
- PMID: 18184807
- PMCID: PMC2206570
- DOI: 10.1073/pnas.0709068105
Preferred side-chain constellations at antiparallel coiled-coil interfaces
Abstract
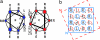
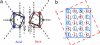
Reliable predictive rules that relate protein sequence to structure would facilitate postgenome predictive biology and the engineering and de novo design of peptides and proteins. Through a combination of experiment and analysis of the protein data bank (PDB), we have deciphered and rationalized new rules for helix-helix interfaces of a common protein-folding and association motif, the antiparallel dimeric coiled coil. These interfaces are defined by a specific pattern of interactions among largely hydrophobic side chains often referred to as knobs-into-holes (KIH) packing: a knob from one helix inserts into a hole formed by four residues on the partner. Previous work has focused on lateral interactions within the KIH motif, for example, between an a position on one helix and a d' position on the other in an antiparallel coiled coil. We show that vertical interactions within the KIH motif, such as a'-a-a', are energetically important as well. The experimental and database analyses concur regarding preferred vertical combinations, which can be rationalized as leading to favorable side-chain interactions that we call constellations. The findings presented here highlight an unanticipated level of complexity in coiled-coil interactions, and our analysis of a few specific constellations illustrates a general, multipronged approach to addressing this complexity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The d'--d--d' vertical triad is less discriminating than the a'--a--a' vertical triad in the antiparallel coiled-coil dimer motif.J Am Chem Soc. 2012 Feb 8;134(5):2626-33. doi: 10.1021/ja208855x. Epub 2012 Jan 31. J Am Chem Soc. 2012. PMID: 22296518 Free PMC article.
-
Strong contributions from vertical triads to helix-partner preferences in parallel coiled coils.J Am Chem Soc. 2012 Sep 26;134(38):15652-5. doi: 10.1021/ja3063088. Epub 2012 Sep 13. J Am Chem Soc. 2012. PMID: 22974448 Free PMC article.
-
Socket: a program for identifying and analysing coiled-coil motifs within protein structures.J Mol Biol. 2001 Apr 13;307(5):1427-50. doi: 10.1006/jmbi.2001.4545. J Mol Biol. 2001. PMID: 11292353
-
The design of coiled-coil structures and assemblies.Adv Protein Chem. 2005;70:79-112. doi: 10.1016/S0065-3233(05)70004-8. Adv Protein Chem. 2005. PMID: 15837514 Review.
-
Structural specificity in coiled-coil interactions.Curr Opin Struct Biol. 2008 Aug;18(4):477-83. doi: 10.1016/j.sbi.2008.04.008. Epub 2008 Jun 12. Curr Opin Struct Biol. 2008. PMID: 18555680 Free PMC article. Review.
Cited by
-
Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.Protein Sci. 2012 Nov;21(11):1705-15. doi: 10.1002/pro.2150. Epub 2012 Oct 1. Protein Sci. 2012. PMID: 22930450 Free PMC article.
-
A set of computationally designed orthogonal antiparallel homodimers that expands the synthetic coiled-coil toolkit.J Am Chem Soc. 2014 Nov 26;136(47):16544-56. doi: 10.1021/ja507847t. Epub 2014 Nov 13. J Am Chem Soc. 2014. PMID: 25337788 Free PMC article.
-
Design of protein-interaction specificity gives selective bZIP-binding peptides.Nature. 2009 Apr 16;458(7240):859-64. doi: 10.1038/nature07885. Nature. 2009. PMID: 19370028 Free PMC article.
-
In situ monitoring of backbone thioester exchange by 19F NMR.Chembiochem. 2009 Sep 4;10(13):2177-81. doi: 10.1002/cbic.200900380. Chembiochem. 2009. PMID: 19644993 Free PMC article. No abstract available.
-
The d'--d--d' vertical triad is less discriminating than the a'--a--a' vertical triad in the antiparallel coiled-coil dimer motif.J Am Chem Soc. 2012 Feb 8;134(5):2626-33. doi: 10.1021/ja208855x. Epub 2012 Jan 31. J Am Chem Soc. 2012. PMID: 22296518 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

