Which missing value imputation method to use in expression profiles: a comparative study and two selection schemes
- PMID: 18186917
- PMCID: PMC2253514
- DOI: 10.1186/1471-2105-9-12
Which missing value imputation method to use in expression profiles: a comparative study and two selection schemes
Abstract
Background: Gene expression data frequently contain missing values, however, most down-stream analyses for microarray experiments require complete data. In the literature many methods have been proposed to estimate missing values via information of the correlation patterns within the gene expression matrix. Each method has its own advantages, but the specific conditions for which each method is preferred remains largely unclear. In this report we describe an extensive evaluation of eight current imputation methods on multiple types of microarray experiments, including time series, multiple exposures, and multiple exposures x time series data. We then introduce two complementary selection schemes for determining the most appropriate imputation method for any given data set.
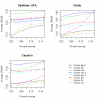
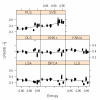
Results: We found that the optimal imputation algorithms (LSA, LLS, and BPCA) are all highly competitive with each other, and that no method is uniformly superior in all the data sets we examined. The success of each method can also depend on the underlying "complexity" of the expression data, where we take complexity to indicate the difficulty in mapping the gene expression matrix to a lower-dimensional subspace. We developed an entropy measure to quantify the complexity of expression matrixes and found that, by incorporating this information, the entropy-based selection (EBS) scheme is useful for selecting an appropriate imputation algorithm. We further propose a simulation-based self-training selection (STS) scheme. This technique has been used previously for microarray data imputation, but for different purposes. The scheme selects the optimal or near-optimal method with high accuracy but at an increased computational cost.
Conclusion: Our findings provide insight into the problem of which imputation method is optimal for a given data set. Three top-performing methods (LSA, LLS and BPCA) are competitive with each other. Global-based imputation methods (PLS, SVD, BPCA) performed better on mcroarray data with lower complexity, while neighbour-based methods (KNN, OLS, LSA, LLS) performed better in data with higher complexity. We also found that the EBS and STS schemes serve as complementary and effective tools for selecting the optimal imputation algorithm.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

