Genetic interactions: the missing links for a better understanding of cancer susceptibility, progression and treatment
- PMID: 18186929
- PMCID: PMC2244643
- DOI: 10.1186/1476-4598-7-4
Genetic interactions: the missing links for a better understanding of cancer susceptibility, progression and treatment
Abstract
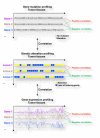
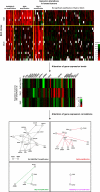
It is increasingly clear that complex networks of relationships between genes and/or proteins govern neoplastic processes. Our understanding of these networks is expanded by the use of functional genomic and proteomic approaches in addition to computational modeling. Concurrently, whole-genome association scans and mutational screens of cancer genomes identify novel cancer genes. Together, these analyses have vastly increased our knowledge of cancer, in terms of both "part lists" and their functional associations. However, genetic interactions have hitherto only been studied in depth in model organisms and remain largely unknown for human systems. Here, we discuss the importance and potential benefits of identifying genetic interactions at the human genome level for creating a better understanding of cancer susceptibility and progression and developing novel effective anticancer therapies. We examine gene expression profiles in the presence and absence of co-amplification of the 8q24 and 20q13 chromosomal regions in breast tumors to illustrate the molecular consequences and complexity of genetic interactions and their role in tumorigenesis. Finally, we highlight current strategies for targeting tumor dependencies and outline potential matrix screening designs for uncovering molecular vulnerabilities in cancer cells.
Figures

References
-
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. - DOI - PubMed
-
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. - DOI - PubMed

