Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes
- PMID: 18187510
- PMCID: PMC2275096
- DOI: 10.1093/nar/gkm1138
Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes
Abstract
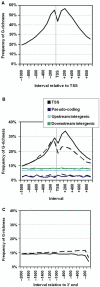
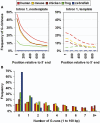
To understand how potential for G-quadruplex formation might influence regulation of gene expression, we examined the 2 kb spanning the transcription start sites (TSS) of the 18 217 human RefSeq genes, distinguishing contributions of template and nontemplate strands. Regions both upstream and downstream of the TSS are G-rich, but the downstream region displays a clear bias toward G-richness on the nontemplate strand. Upstream of the TSS, much of the G-richness and potential for G-quadruplex formation derives from the presence of well-defined canonical regulatory motifs in duplex DNA, including CpG dinucleotides which are sites of regulatory methylation, and motifs recognized by the transcription factor SP1. This challenges the notion that quadruplex formation upstream of the TSS contributes to regulation of gene expression. Downstream of the TSS, G-richness is concentrated in the first intron, and on the nontemplate strand, where polymorphic sequence elements with potential to form G-quadruplex structures and which cannot be accounted for by known regulatory motifs are found in almost 3000 (16%) of the human RefSeq genes, and are conserved through frogs. These elements could in principle be recognized either as DNA or as RNA, providing structural targets for regulation at the level of transcription or RNA processing.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

