Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II
- PMID: 18187511
- PMCID: PMC2275077
- DOI: 10.1093/nar/gkm1068
Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II
Abstract
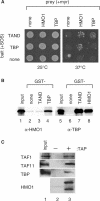
Saccharomyces cerevisiae HMO1, a high mobility group B (HMGB) protein, associates with the rRNA locus and with the promoters of many ribosomal protein genes (RPGs). Here, the Sos recruitment system was used to show that HMO1 interacts with TBP and the N-terminal domain (TAND) of TAF1, which are integral components of TFIID. Biochemical studies revealed that HMO1 copurifies with TFIID and directly interacts with TBP but not with TAND. Deletion of HMO1 (Deltahmo1) causes a severe cold-sensitive growth defect and decreases transcription of some TAND-dependent genes. Deltahmo1 also affects TFIID occupancy at some RPG promoters in a promoter-specific manner. Interestingly, over-expression of HMO1 delays colony formation of taf1 mutants lacking TAND (taf1DeltaTAND), but not of the wild-type strain, indicating a functional link between HMO1 and TAND. Furthermore, Deltahmo1 exhibits synthetic growth defects in some spt15 (TBP) and toa1 (TFIIA) mutants while it rescues growth defects of some sua7 (TFIIB) mutants. Importantly, Deltahmo1 causes an upstream shift in transcriptional start sites of RPS5, RPS16A, RPL23B, RPL27B and RPL32, but not of RPS31, RPL10, TEF2 and ADH1, indicating that HMO1 may participate in start site selection of a subset of class II genes presumably via its interaction with TFIID.
Figures






Similar articles
-
Hmo1 directs pre-initiation complex assembly to an appropriate site on its target gene promoters by masking a nucleosome-free region.Nucleic Acids Res. 2011 May;39(10):4136-50. doi: 10.1093/nar/gkq1334. Epub 2011 Feb 2. Nucleic Acids Res. 2011. PMID: 21288884 Free PMC article.
-
Crystal structure, biochemical and genetic characterization of yeast and E. cuniculi TAF(II)5 N-terminal domain: implications for TFIID assembly.J Mol Biol. 2007 May 18;368(5):1292-306. doi: 10.1016/j.jmb.2007.02.039. Epub 2007 Feb 22. J Mol Biol. 2007. PMID: 17397863
-
Both HMG boxes in Hmo1 are essential for DNA binding in vitro and in vivo.Biosci Biotechnol Biochem. 2015;79(3):384-93. doi: 10.1080/09168451.2014.978258. Epub 2014 Nov 20. Biosci Biotechnol Biochem. 2015. PMID: 25410521
-
Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
-
Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review.
Cited by
-
A Random Screen Using a Novel Reporter Assay System Reveals a Set of Sequences That Are Preferred as the TATA or TATA-Like Elements in the CYC1 Promoter of Saccharomyces cerevisiae.PLoS One. 2015 Jun 5;10(6):e0129357. doi: 10.1371/journal.pone.0129357. eCollection 2015. PLoS One. 2015. PMID: 26046838 Free PMC article.
-
Hmo1: A versatile member of the high mobility group box family of chromosomal architecture proteins.World J Biol Chem. 2024 Aug 12;15(1):97938. doi: 10.4331/wjbc.v15.i1.97938. World J Biol Chem. 2024. PMID: 39156122 Free PMC article. Review.
-
Yeast Crf1p: An activator in need is an activator indeed.Comput Struct Biotechnol J. 2021 Dec 8;20:107-116. doi: 10.1016/j.csbj.2021.12.003. eCollection 2022. Comput Struct Biotechnol J. 2021. PMID: 34976315 Free PMC article. Review.
-
Identification of a transcriptional activation domain in yeast repressor activator protein 1 (Rap1) using an altered DNA-binding specificity variant.J Biol Chem. 2017 Apr 7;292(14):5705-5723. doi: 10.1074/jbc.M117.779181. Epub 2017 Feb 14. J Biol Chem. 2017. PMID: 28196871 Free PMC article.
-
Coordination of Ribosomal Protein and Ribosomal RNA Gene Expression in Response to TOR Signaling.Curr Genomics. 2009 May;10(3):198-205. doi: 10.2174/138920209788185261. Curr Genomics. 2009. PMID: 19881913 Free PMC article.
References
-
- Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. - PubMed
-
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. - PubMed
-
- Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 2005;30:275–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

