Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II
- PMID: 18187511
- PMCID: PMC2275077
- DOI: 10.1093/nar/gkm1068
Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II
Abstract
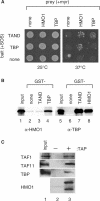
Saccharomyces cerevisiae HMO1, a high mobility group B (HMGB) protein, associates with the rRNA locus and with the promoters of many ribosomal protein genes (RPGs). Here, the Sos recruitment system was used to show that HMO1 interacts with TBP and the N-terminal domain (TAND) of TAF1, which are integral components of TFIID. Biochemical studies revealed that HMO1 copurifies with TFIID and directly interacts with TBP but not with TAND. Deletion of HMO1 (Deltahmo1) causes a severe cold-sensitive growth defect and decreases transcription of some TAND-dependent genes. Deltahmo1 also affects TFIID occupancy at some RPG promoters in a promoter-specific manner. Interestingly, over-expression of HMO1 delays colony formation of taf1 mutants lacking TAND (taf1DeltaTAND), but not of the wild-type strain, indicating a functional link between HMO1 and TAND. Furthermore, Deltahmo1 exhibits synthetic growth defects in some spt15 (TBP) and toa1 (TFIIA) mutants while it rescues growth defects of some sua7 (TFIIB) mutants. Importantly, Deltahmo1 causes an upstream shift in transcriptional start sites of RPS5, RPS16A, RPL23B, RPL27B and RPL32, but not of RPS31, RPL10, TEF2 and ADH1, indicating that HMO1 may participate in start site selection of a subset of class II genes presumably via its interaction with TFIID.
Figures






References
-
- Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. - PubMed
-
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. - PubMed
-
- Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 2005;30:275–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

