SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system
- PMID: 18188155
- PMCID: PMC2234343
- DOI: 10.1038/sj.emboj.7601966
SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system
Abstract
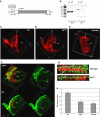
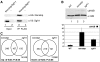
Nonstop, which has previously been shown to have homology to ubiquitin proteases, is required for proper termination of axons R1-R6 in the optic lobe of the developing Drosophila eye. Herein, we establish that Nonstop actually functions as an ubiquitin protease to control the levels of ubiquitinated histone H2B in flies. We further establish that Nonstop is the functional homolog of yeast Ubp8, and can substitute for Ubp8 function in yeast cells. In yeast, Ubp8 activity requires Sgf11. We show that in Drosophila, loss of Sgf11 function causes similar photoreceptor axon-targeting defects as loss of Nonstop. Ubp8 and Sgf11 are components of the yeast SAGA complex, suggesting that Nonstop function might be mediated through the Drosophila SAGA complex. Indeed, we find that Nonstop does associate with SAGA components in flies, and mutants in other SAGA subunits display nonstop phenotypes, indicating that SAGA complex is required for accurate axon guidance in the optic lobe. Candidate genes regulated by SAGA that may be required for correct axon targeting were identified by microarray analysis of gene expression in SAGA mutants.
Figures




References
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 - PubMed
-
- Awasaki T, Ito K (2004) Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol 14: 668–677 - PubMed
-
- Brennan CA, Li TR, Bender M, Hsiung F, Moses K (2001) Broad-complex, but not ecdysone receptor, is required for progression of the morphogenetic furrow in the Drosophila eye. Development 128: 1–11 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

