Genetic analysis of hantaviruses carried by Myodes and Microtus rodents in Buryatia
- PMID: 18190679
- PMCID: PMC2248171
- DOI: 10.1186/1743-422X-5-4
Genetic analysis of hantaviruses carried by Myodes and Microtus rodents in Buryatia
Abstract
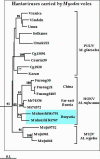
Hantavirus genome sequences were recovered from tissue samples of Myodes rufocanus, Microtus fortis and Microtus oeconomus captured in the Baikal area of Buryatia, Russian Federation. Genetic analysis of S- and M-segment sequences of Buryatian hantavirus strains showed that Myodes-associated strains belong to Hokkaido virus (HOKV) type while Microtus-associated strains belong to Vladivostok virus (VLAV) type. On phylogenetic trees Buryatian HOKV strains were clustered together with M. rufocanus- originated strains from Japan, China and Far-East Russia (Primorsky region). Buryatian Microtus- originated strains shared a common recent ancestor with M. fortis- originated VLAV strain from Far-East Russia (Vladivostok area). Our data (i) confirm that M. rufocanus carries a hantavirus which is similar to but distinct from both Puumala virus carried by M. glareolus and Muju virus associated with M. regulus, (ii) confirm that M. fortis is the natural host for VLAV, and (iii) suggest M. oeconomus as an alternative host for VLAV.
Figures

Similar articles
-
Isolation and genetic characterization of hantaviruses carried by Microtus voles in China.J Med Virol. 2008 Apr;80(4):680-8. doi: 10.1002/jmv.21119. J Med Virol. 2008. PMID: 18297708
-
Genetic diversities of hantaviruses among rodents in Hokkaido, Japan and Far East Russia.Virus Res. 1999 Feb;59(2):219-28. doi: 10.1016/s0168-1702(98)00141-5. Virus Res. 1999. PMID: 10082393
-
Genetic analysis of hantaviruses carried by reed voles Microtus fortis in China.Virus Res. 2008 Oct;137(1):122-8. doi: 10.1016/j.virusres.2008.06.012. Epub 2008 Aug 3. Virus Res. 2008. PMID: 18644410
-
A comparative epidemiological study of hantavirus infection in Japan and Far East Russia.Jpn J Vet Res. 2007 Feb;54(4):145-61. Jpn J Vet Res. 2007. PMID: 17405352 Review.
-
Hantaviruses in Africa.Virus Res. 2014 Jul 17;187:34-42. doi: 10.1016/j.virusres.2013.12.039. Epub 2014 Jan 7. Virus Res. 2014. PMID: 24406800 Review.
Cited by
-
Hokkaido genotype of Puumala virus in the grey red-backed vole (Myodes rufocanus) and northern red-backed vole (Myodes rutilus) in Siberia.Infect Genet Evol. 2015 Jul;33:304-13. doi: 10.1016/j.meegid.2015.05.021. Epub 2015 May 21. Infect Genet Evol. 2015. PMID: 26003760 Free PMC article.
-
Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea).PLoS One. 2009 Jul 7;4(7):e6149. doi: 10.1371/journal.pone.0006149. PLoS One. 2009. PMID: 19582155 Free PMC article.
-
Phylogenetic analysis of Puumala virus subtype Bavaria, characterization and diagnostic use of its recombinant nucleocapsid protein.Virus Genes. 2011 Oct;43(2):177-91. doi: 10.1007/s11262-011-0620-x. Epub 2011 May 20. Virus Genes. 2011. PMID: 21598005
-
Phylogeography of Puumala orthohantavirus in Europe.Viruses. 2019 Jul 24;11(8):679. doi: 10.3390/v11080679. Viruses. 2019. PMID: 31344894 Free PMC article.
-
Global Diversity and Distribution of Hantaviruses and Their Hosts.Ecohealth. 2018 Mar;15(1):163-208. doi: 10.1007/s10393-017-1305-2. Epub 2018 Apr 30. Ecohealth. 2018. PMID: 29713899 Review.
References
-
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Tesh RB. Bunyaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. Virus taxonomy VIIIth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier Academic Press; 2005. pp. 695–716.
-
- Lundkvist Å, Plyusnin A. Molecular epidemiology of hantavirus infections. In: Leitner T, editor. The Molecular Epidemiology of Human Viruses. Kluwer Academic Publishers; 2002. pp. 351–384.
-
- Kariwa H, Yoshimatsu K, Araki K, Chayama K, Kumada H, Ogino M, Ebihara H, Murphy ME, Mizutani T, Takashima I, Arikawa J. Detection of hantaviral antibodies among patients with hepatitis of unknown etiology in Japan. Microbiol and Immunol. 2000;44:357–362. - PubMed
-
- Vapalahti O, Lundkvist Å, Fedorov V, Conroy C, Hirvonen S, Plyusnina A, Nemirov K, Fredga K, Cook J, Niemimaa J, Kaikusalo A, Henttonen H, Vaheri A, Plyusnin A. Isolation and characterization of a hantavirus from Lemmus sibiricus: evidence for host-switch during hantavirus evolution. J Virol. 1999;73:5586–5592. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

