Maintenance forced by a restriction-modification system can be modulated by a region in its modification enzyme not essential for methyltransferase activity
- PMID: 18192396
- PMCID: PMC2258900
- DOI: 10.1128/JB.01319-07
Maintenance forced by a restriction-modification system can be modulated by a region in its modification enzyme not essential for methyltransferase activity
Abstract
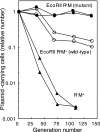
Several type II restriction-modification gene complexes can force their maintenance on their host bacteria by killing cells that have lost them in a process called postsegregational killing or genetic addiction. It is likely to proceed by dilution of the modification enzyme molecule during rounds of cell division following the gene loss, which exposes unmethylated recognition sites on the newly replicated chromosomes to lethal attack by the remaining restriction enzyme molecules. This process is in apparent contrast to the process of the classical types of postsegregational killing systems, in which built-in metabolic instability of the antitoxin allows release of the toxin for lethal action after the gene loss. In the present study, we characterize a mutant form of the EcoRII gene complex that shows stronger capacity in such maintenance. This phenotype is conferred by an L80P amino acid substitution (T239C nucleotide substitution) mutation in the modification enzyme. This mutant enzyme showed decreased DNA methyltransferase activity at a higher temperature in vivo and in vitro than the nonmutated enzyme, although a deletion mutant lacking the N-terminal 83 amino acids did not lose activity at either of the temperatures tested. Under a condition of inhibited protein synthesis, the activity of the L80P mutant was completely lost at a high temperature. In parallel, the L80P mutant protein disappeared more rapidly than the wild-type protein. These results demonstrate that the capability of a restriction-modification system in forcing maintenance on its host can be modulated by a region of its antitoxin, the modification enzyme, as in the classical postsegregational killing systems.
Figures







References
-
- Afif, H., N. Allali, M. Couturier, and L. Van Melderen. 2001. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 4173-82. - PubMed
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. - PubMed
-
- Bachmann, B. J. 1987. Derivation and genotypes of some mutant derivatives of Escherichia coli K-12, p. 1190-1219. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology, vol. 2. American Society for Microbiology, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

