Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example
- PMID: 18192413
- PMCID: PMC2258636
- DOI: 10.1128/AEM.02403-07
Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example
Abstract
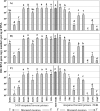
We investigated the effects of internal primer-template mismatches on the efficiency of PCR amplification using the 16S rRNA gene as the model template DNA. We observed that the presence of a single mismatch in the second half of the primer extension sequence can result in an underestimation of up to 1,000-fold of the gene copy number, depending on the primer and position of the mismatch.
Figures

References
-
- Ayyadevara, S., J. J. Thaden, and R. J. S. Reis. 2000. Discrimination of primer 3′-nucleotide mismatch by Taq DNA polymerase during polymerase chain reaction. Anal. Biochem. 284:11-18. - PubMed
-
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. - PubMed
-
- Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

