Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome
- PMID: 18193060
- PMCID: PMC2546704
- DOI: 10.1038/nsmb.1375
Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome
Abstract
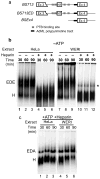
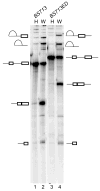
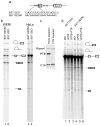
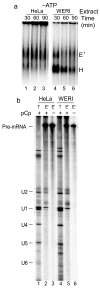
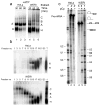
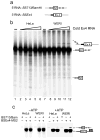
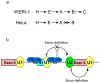
The polypyrimidine tract binding protein (PTB) binds pre-mRNAs to alter splice-site choice. We characterized a series of spliceosomal complexes that assemble on a pre-mRNA under conditions of either PTB-mediated splicing repression or its absence. In the absence of repression, exon definition complexes that were assembled downstream of the regulated exon could progress to pre-spliceosomal A complexes and functional spliceosomes. Under PTB-mediated repression, assembly was arrested at an A-like complex that was unable to transition to spliceosomal complexes. Trans-splicing experiments indicated that, even when the U1 and U2 small nuclear ribonucleoprotein particles (snRNPs) are properly bound to the upstream and downstream exons, the presence of PTB prevents the interaction of the two exon complexes. Proteomic analyses of these complexes provide a new description of exon definition complexes, and indicate that splicing regulators can act on the transition between the exon definition complex and an intron-defined spliceosome.
Figures

Similar articles
-
Structural insights into the cross-exon to cross-intron spliceosome switch.Nature. 2024 Jun;630(8018):1012-1019. doi: 10.1038/s41586-024-07458-1. Epub 2024 May 22. Nature. 2024. PMID: 38778104 Free PMC article.
-
Polypyrimidine tract binding protein blocks the 5' splice site-dependent assembly of U2AF and the prespliceosomal E complex.Mol Cell. 2005 Aug 19;19(4):485-96. doi: 10.1016/j.molcel.2005.07.014. Mol Cell. 2005. PMID: 16109373 Free PMC article.
-
A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart.RNA. 1997 Sep;3(9):996-1015. RNA. 1997. PMID: 9292499 Free PMC article.
-
Pre-mRNA splicing: a complex picture in higher definition.Trends Biochem Sci. 2008 Jun;33(6):243-6. doi: 10.1016/j.tibs.2008.04.004. Epub 2008 May 9. Trends Biochem Sci. 2008. PMID: 18472266 Review.
-
Evolution of the Early Spliceosomal Complex-From Constitutive to Regulated Splicing.Int J Mol Sci. 2021 Nov 18;22(22):12444. doi: 10.3390/ijms222212444. Int J Mol Sci. 2021. PMID: 34830325 Free PMC article. Review.
Cited by
-
Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins.Biomolecules. 2015 Jul 15;5(3):1441-66. doi: 10.3390/biom5031441. Biomolecules. 2015. PMID: 26184334 Free PMC article.
-
A day in the life of the spliceosome.Nat Rev Mol Cell Biol. 2014 Feb;15(2):108-21. doi: 10.1038/nrm3742. Nat Rev Mol Cell Biol. 2014. PMID: 24452469 Free PMC article. Review.
-
The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex.Genes Dev. 2011 May 1;25(9):972-83. doi: 10.1101/gad.2038011. Genes Dev. 2011. PMID: 21536736 Free PMC article.
-
A U1-U2 snRNP interaction network during intron definition.Mol Cell Biol. 2012 Jan;32(2):470-8. doi: 10.1128/MCB.06234-11. Epub 2011 Nov 7. Mol Cell Biol. 2012. PMID: 22064476 Free PMC article.
-
Large-scale remodeling of a repressed exon ribonucleoprotein to an exon definition complex active for splicing.Elife. 2016 Nov 24;5:e19743. doi: 10.7554/eLife.19743. Elife. 2016. PMID: 27882870 Free PMC article.
References
-
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–8. - PubMed
-
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007 - PubMed
-
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–38. - PubMed
-
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nature. 2005;6:386–398. - PubMed
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

