Structural basis of the metal specificity for nickel regulatory protein NikR
- PMID: 18193897
- PMCID: PMC5914170
- DOI: 10.1021/bi702006h
Structural basis of the metal specificity for nickel regulatory protein NikR
Abstract
In the presence of excess nickel, Escherichia coli NikR regulates cellular nickel uptake by suppressing the transcription of the nik operon, which encodes the nickel uptake transporter, NikABCDE. Previously published in vitro studies have shown that NikR is capable of binding a range of divalent transition metal ions in addition to Ni2+, including Co2+, Cu2+, Zn2+, and Cd2+. To understand how the high-affinity nickel binding site of NikR is able to accommodate these other metal ions, and to improve our understanding of NikR's mechanism of binding to DNA, we have determined structures of the metal-binding domain (MBD) of NikR in the apo form and in complex with Cu2+ and Zn2+ ions and compared them with the previously published structures with Ni2+. We observe that Cu2+ ions bind in a manner very similar to that of Ni2+, with a square planar geometry but with longer bond lengths. Crystals grown in the presence of Zn2+ reveal a protein structure similar to that of apo MBD with a disordered alpha3 helix, but with two electron density peaks near the Ni2+ binding site corresponding to two Zn2+ ions. These structural findings along with biochemical data on NikR support a hypothesis that ordering of the alpha3 helix is important for repressor activation.
Figures

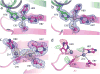

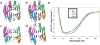
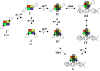
Similar articles
-
Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.Biochemistry. 2010 Sep 14;49(36):7830-8. doi: 10.1021/bi100923j. Biochemistry. 2010. PMID: 20704276 Free PMC article.
-
Metal-selective DNA-binding response of Escherichia coli NikR.Biochemistry. 2004 Aug 10;43(31):10029-38. doi: 10.1021/bi049404k. Biochemistry. 2004. PMID: 15287730
-
High-affinity Ni2+ binding selectively promotes binding of Helicobacter pylori NikR to its target urease promoter.J Mol Biol. 2008 Nov 28;383(5):1129-43. doi: 10.1016/j.jmb.2008.08.066. Epub 2008 Sep 4. J Mol Biol. 2008. PMID: 18790698
-
Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators.Biometals. 2005 Aug;18(4):413-28. doi: 10.1007/s10534-005-3716-8. Biometals. 2005. PMID: 16158234 Review.
-
Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors.Nat Prod Rep. 2010 May;27(5):658-67. doi: 10.1039/b906683g. Epub 2010 Mar 5. Nat Prod Rep. 2010. PMID: 20442957 Review.
Cited by
-
Nickel-based Enzyme Systems.J Biol Chem. 2009 Jul 10;284(28):18571-5. doi: 10.1074/jbc.R900020200. Epub 2009 Apr 10. J Biol Chem. 2009. PMID: 19363030 Free PMC article. Review.
-
Bacterial Metallostasis: Metal Sensing, Metalloproteome Remodeling, and Metal Trafficking.Chem Rev. 2024 Dec 25;124(24):13574-13659. doi: 10.1021/acs.chemrev.4c00264. Epub 2024 Dec 10. Chem Rev. 2024. PMID: 39658019 Free PMC article. Review.
-
Holo-Ni(II)HpNikR is an asymmetric tetramer containing two different nickel-binding sites.J Am Chem Soc. 2010 Oct 20;132(41):14447-56. doi: 10.1021/ja104118r. J Am Chem Soc. 2010. PMID: 20863122 Free PMC article.
-
Mechanistic insights into the nickel-dependent allosteric response of the Helicobacter pylori NikR transcription factor.J Biol Chem. 2023 Jan;299(1):102785. doi: 10.1016/j.jbc.2022.102785. Epub 2022 Dec 9. J Biol Chem. 2023. PMID: 36502919 Free PMC article.
-
Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.Biochemistry. 2010 Sep 14;49(36):7830-8. doi: 10.1021/bi100923j. Biochemistry. 2010. PMID: 20704276 Free PMC article.
References
-
- Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. - PubMed
-
- Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27:239–261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

