Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies
- PMID: 18194512
- PMCID: PMC2262094
- DOI: 10.1186/1471-2199-9-3
Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies
Abstract
Background: Due to the limited RNA amounts from endomyocardial biopsies (EMBs) and low expression levels of certain genes, gene expression analyses by conventional real-time RT-PCR are restrained in EMBs. We applied two preamplification techniques, the TaqMan(R) PreAmp Master Mix (T-PreAmp) and a multiplex preamplification following a sequence specific reverse transcription (SSRT-PreAmp).
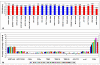
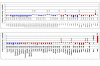
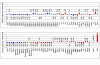
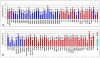
Results: T-PreAmp encompassing 92 gene assays with 14 cycles resulted in a mean improvement of 7.24 +/- 0.33 Ct values. The coefficients for inter- (1.89 +/- 0.48%) and intra-assay variation (0.85 +/- 0.45%) were low for all gene assays tested (<4%). The PreAmp uniformity values related to the reference gene CDKN1B for 91 of the investigated gene assays (except for CD56) were -0.38 +/- 0.33, without significant differences between self-designed and ABI inventoried Taqman(R) gene assays. Only two of the tested Taqman(R) ABI inventoried gene assays (HPRT-ABI and CD56) did not maintain PreAmp uniformity levels between -1.5 and +1.5. In comparison, the SSRT-PreAmp tested on 8 self-designed gene assays yielded higher Ct improvement (9.76 +/- 2.45), however was not as robust regarding the maintenance of PreAmp uniformity related to HPRT-CCM (-3.29 +/- 2.40; p < 0.0001), and demonstrated comparable intra-assay CVs (1.47 +/- 0.74), albeit higher inter-assay CVs (5.38 +/- 2.06; p = 0.01). Comparing EMBs from each 10 patients with dilated cardiomyopathy (DCM) and inflammatory cardiomyopathy (DCMi), T-PreAmp real-time RT-PCR analyses revealed differential regulation regarding 27 (30%) of the investigated 90 genes related to both HPRT-CCM and CDKN1B. Ct values of HPRT and CDKN1B did not differ in equal RNA amounts from explanted DCM and donor hearts.
Conclusion: In comparison to the SSRT-PreAmp, T-PreAmp enables a relatively simple workflow, and results in a robust PreAmp of multiple target genes (at least 92 gene assays as tested here) by a mean Ct improvement around 7 cycles, and in a lower inter-assay variance in RNA derived from EMBs. Preliminary analyses comparing EMBs from DCM and DCMi patients, revealing differential regulation regarding 30% of the investigated genes, confirm that T-PreAmp is a suitable tool to perform gene expression analyses in EMBs, expanding gene expression investigations with the limited RNA/cDNA amounts derived from EMBs. CDKN1B, in addition to its function as a reference gene for the calculation of PreAmp uniformity, might serve as a suitable housekeeping gene for real-time RT-PCR analyses of myocardial tissues.
Figures








Similar articles
-
Real-time RT-PCR for the detection of beta-adrenoceptor messenger RNAs in small human endomyocardial biopsies.J Mol Cell Cardiol. 2001 Dec;33(12):2121-33. doi: 10.1006/jmcc.2001.1475. J Mol Cell Cardiol. 2001. PMID: 11735259
-
Expression of functional T-cell markers and T-cell receptor Vbeta repertoire in endomyocardial biopsies from patients presenting with acute myocarditis and dilated cardiomyopathy.Eur J Heart Fail. 2011 Jun;13(6):611-8. doi: 10.1093/eurjhf/hfr014. Epub 2011 Mar 19. Eur J Heart Fail. 2011. PMID: 21422001
-
Reliability and reproducibility of a RNA preamplification method for low-density array analysis from formalin-fixed paraffin-embedded breast cancer samples.Diagn Mol Pathol. 2009 Jun;18(2):112-8. doi: 10.1097/PDM.0b013e3181831320. Diagn Mol Pathol. 2009. PMID: 19430292
-
In-house HIV-1 RNA real-time RT-PCR assays: principle, available tests and usefulness in developing countries.Expert Rev Mol Diagn. 2008 Sep;8(5):635-50. doi: 10.1586/14737159.8.5.635. Expert Rev Mol Diagn. 2008. PMID: 18785811 Review.
-
[RT-PCR in clinical diagnosis].Ann Biol Clin (Paris). 2003 Nov-Dec;61(6):635-44. Ann Biol Clin (Paris). 2003. PMID: 14711604 Review. French.
Cited by
-
Chronic psychosocial stressors and salivary biomarkers in emerging adults.Psychoneuroendocrinology. 2012 Aug;37(8):1158-70. doi: 10.1016/j.psyneuen.2011.11.010. Epub 2011 Dec 14. Psychoneuroendocrinology. 2012. PMID: 22172638 Free PMC article. Clinical Trial.
-
Loss of CDKN2A and CDKN2B expression is associated with disease recurrence in oral cancer.J Oral Maxillofac Pathol. 2019 Jan-Apr;23(1):82-89. doi: 10.4103/jomfp.JOMFP_184_18. J Oral Maxillofac Pathol. 2019. PMID: 31110422 Free PMC article.
-
Biovigilance for the Quality and Safety of Medical Products of Human Origin.J Clin Med Res. 2018 Dec;10(12):857-867. doi: 10.14740/jocmr3549w. Epub 2018 Oct 30. J Clin Med Res. 2018. PMID: 30425757 Free PMC article. Review.
-
Selection of reference genes for quantitative real time PCR (qPCR) assays in tissue from human ascending aorta.PLoS One. 2014 May 19;9(5):e97449. doi: 10.1371/journal.pone.0097449. eCollection 2014. PLoS One. 2014. PMID: 24841551 Free PMC article.
-
Changes of microRNAs-192, 196a and 203 correlate with Barrett's esophagus diagnosis and its progression compared to normal healthy individuals.Diagn Pathol. 2011 Nov 17;6:114. doi: 10.1186/1746-1596-6-114. Diagn Pathol. 2011. PMID: 22094011 Free PMC article.
References
-
- Noutsias M, Seeberg B, Schultheiss HP, Kuhl U. Expression of cell adhesion molecules in dilated cardiomyopathy: evidence for endothelial activation in inflammatory cardiomyopathy. Circulation. 1999;99:2124–2131. - PubMed
-
- Kuhl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, Poller W, Kandolf R, Schultheiss HP. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with "idiopathic" left ventricular dysfunction. Circulation. 2005;111:887–893. doi: 10.1161/01.CIR.0000155616.07901.35. - DOI - PubMed
-
- Staudt A, Schaper F, Stangl V, Plagemann A, Bohm M, Merkel K, Wallukat G, Wernecke KD, Stangl K, Baumann G, et al. Immunohistological changes in dilated cardiomyopathy induced by immunoadsorption therapy and subsequent immunoglobulin substitution. Circulation. 2001;103:2681–2686. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

