Substrate specificity of new methyl-directed DNA endonuclease GlaI
- PMID: 18194583
- PMCID: PMC2257971
- DOI: 10.1186/1471-2199-9-7
Substrate specificity of new methyl-directed DNA endonuclease GlaI
Abstract
Background: Recently, we have discovered site-specific endonucleases, which recognize and cleave only DNA sequences with 5-methylcytosine. Two specificities of such endonucleases have been described. Enzymes BisI, BlsI, and GluI are isoschizomers and hydrolyze the DNA sequence 5'-GCNGC-3'/3'-CGNCG-5', which is methylated in different ways. The enzyme GlaI cleaves the DNA sequence 5'-GCGC-3'/3'-CGCG-5' if there are two, three or four 5-methylcytosines. The goal of the present work is to study in detail the composition of recognition sequence and effect of the methylated cytosines on the efficiency of DNA cleavage by the methyl-directed DNA endonuclease GlaI RESULTS: In a recent work we have studied the dependence of GlaI activity on the quantity and location of 5-methylcytosines in the enzyme recognition sequence 5'-GCGC-3'/3'-CGCG-5'. A significant DNA cleavage has been observed for oligonucleotide duplexes, which include either three or four 5-methylcytosines. In this work we have studied dependence of the GlaI activity on quantity and location of methylated cytosines, as well as on composition of the recognition sequence.
Conclusion: The list of good substrates for GlaI includes a fully methylated site 5'-CGCG-3'/3'-GCGC-5', sites with three cytosines of a general structure 5'-PuMGM-3'/3'-PyGMG-5', and one recognition sequence with two methylated cytosines 5'-AMGT-3'/3'-TGMA-5', where M is 5-methylcytosine.GlaI intermediate substrates include sites with three methylated cytosines of a general structure 5'-GMPuM-3'/3'-MGPyG-5', as well as a site with two methylcytosines 5'-GMGT-3'/3'-CGMA-5'. The site 5'-GMGC-3'/3'-CGMG-5' may be considered a low activity substrate.
Figures
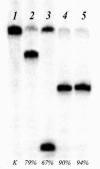
 .
.
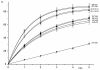
 ; b) G13*/G14 (5'-TTTTC
; b) G13*/G14 (5'-TTTTC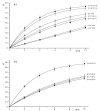
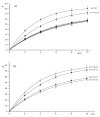
 ; b) G12*/G17 (5'-CGTCA
; b) G12*/G17 (5'-CGTCA
References
-
- Chmuzh EV, Kashirina Yu G, Tomilova Yu E, Mezentseva NV, Dedkov VS, Gonchar DA, Abdurashitov MA, Degtyarev SKh. New endonuclease restriction BisI from Bacillus subtilis T30 recognizes methylated sequence DNA 5'-G(5mC)N^GC-3' http://science.sibenzyme.com/article8_article_7_1.phtml
-
- Chernukhin VA, Nayakshina TN, Abdurashitov MA, Tomilova Yu E, Mezentseva NV, Dedkov VS, Mikhnenkova NA, Gonchar DA, Degtyarev SKh. New restriction endonuclease GlaI recognizes methylatedsequence5'-G(5mC)G^C-3' http://science.sibenzyme.com/article8_article_11_1.phtml
-
- Chernukhin VA, Tomilova Yu E, Chmuzh EV, Sokolova OO, Dedkov VS, Degtyarev SKh. Site-specific endonuclease BlsI recognizes DNA sequence 5'-G(5mC)NG^C-3' and cleaves it producing3'stickyends http://science.sibenzyme.com/article8_article_25_1.phtml
-
- Chernukhin VA, Chmuzh EV, Tomilova Yu E, Nayakshina TN, Gonchar DA, Dedkov VS, Degtyarev SKh. A novel site-specific endonuclease GluI recognizes methylated DNA sequence 5'-G(5mC)N^G(5mC)-3'/3'-(5mC)GN^(5mC)G http://science.sibenzyme.com/article8_article_24_1.phtml
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

