Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium
- PMID: 18195033
- PMCID: PMC2258808
- DOI: 10.1128/IAI.01383-07
Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium
Abstract
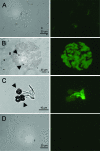
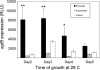
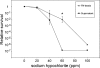
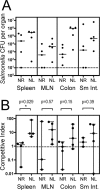
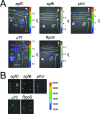
The Salmonella rdar (red, dry, and rough) morphotype is an aggregative and resistant physiology that has been linked to survival in nutrient-limited environments. Growth of Salmonella enterica serovar Typhimurium was analyzed in a variety of nutrient-limiting conditions to determine whether aggregation would occur at low cell densities and whether the rdar morphotype was involved in this process. The resulting cultures consisted of two populations of cells, aggregated and nonaggregated, with the aggregated cells preferentially displaying rdar morphotype gene expression. The two groups of cells could be separated based on the principle that aggregated cells were producing greater amounts of thin aggregative fimbriae (Tafi or curli). In addition, the aggregated cells retained some physiological characteristics of the rdar morphotype, such as increased resistance to sodium hypochlorite. Competitive infection experiments in mice showed that nonaggregative DeltaagfA cells outcompeted rdar-positive wild-type cells in all tissues analyzed, indicating that aggregation via the rdar morphotype was not a virulence adaptation in Salmonella enterica serovar Typhimurium. Furthermore, in vivo imaging experiments showed that Tafi genes were not expressed during infection but were expressed once Salmonella was passed out of the mice into the feces. We hypothesize that the primary role of the rdar morphotype is to enhance Salmonella survival outside the host, thereby aiding in transmission.
Figures


References
-
- Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 62443-2452. - PubMed
-
- Ben Nasr, A., A. Olsen, U. Sjobring, W. Muller-Esterl, and L. Bjorck. 1996. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20927-935. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

