A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation
- PMID: 18196178
- PMCID: PMC2267755
- DOI: 10.1016/j.cpc.2007.08.012
A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation
Abstract
Analytical ultracentrifugation allows one to measure in real-time the concentration gradients arising from the application of a centrifugal force to macromolecular mixtures in solution. In the last decade, the ability to efficiently solve the partial differential equation governing the ultracentrifugal sedimentation and diffusion process, the Lamm equation, has spawned significant progress in the application of sedimentation velocity analytical ultracentrifugation for the study of biological macromolecules, for example, the characterization of protein oligomeric states and the study of reversible multi-protein complexes in solution. The present work describes a numerical algorithm that can provide an improvement in accuracy or efficiency over existing algorithms by more than one order of magnitude, and thereby greatly facilitate the practical application of sedimentation velocity analysis, in particular, for the study of multi-component macromolecular mixtures. It is implemented in the public domain software SEDFIT for the analysis of experimental data.
Figures


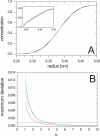

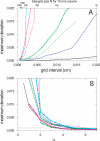

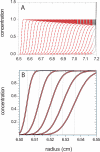
References
-
- Svedberg T, Pedersen KO. The ultracentrifuge. Oxford University Press; London: 1940.
-
- Svedberg T, Fahraeus R. A new method for the determination of the molecular weight of the proteins. J Am Chem Soc. 1926;48:320–438.
-
- Schachman HK. Ultracentrifugation in Biochemistry. Academic Press; New York: 1959.
-
- Signer R, Gross H. Ultrazentrifugale Polydispersitätsbestimmungen an hochpolymeren Stoffen. Helv. Chim. Acta. 1934;17:726.
Grants and funding
LinkOut - more resources
Full Text Sources
